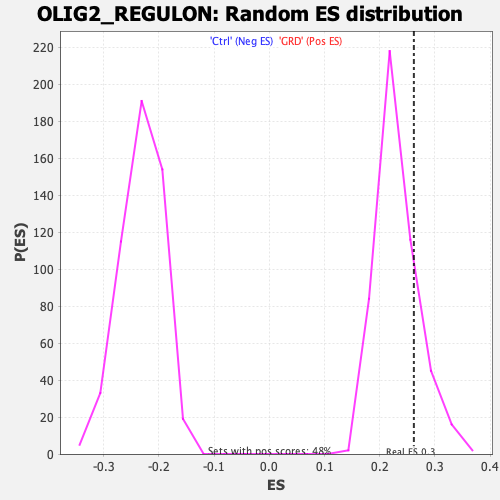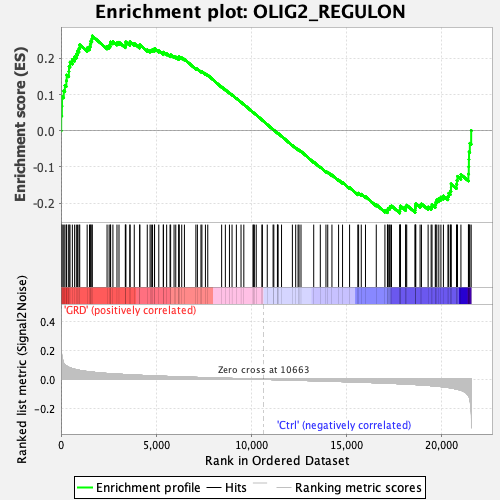
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | GRD |
| GeneSet | OLIG2_REGULON |
| Enrichment Score (ES) | 0.26190338 |
| Normalized Enrichment Score (NES) | 1.1310581 |
| Nominal p-value | 0.19254659 |
| FDR q-value | 0.24532385 |
| FWER p-Value | 0.966 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FSTL1 | FSTL1 | 10 | 0.234 | 0.0418 | Yes |
| 2 | FEZ1 | FEZ1 | 46 | 0.154 | 0.0681 | Yes |
| 3 | AGPAT5 | AGPAT5 | 54 | 0.148 | 0.0946 | Yes |
| 4 | MAP2 | MAP2 | 145 | 0.108 | 0.1100 | Yes |
| 5 | TMPRSS5 | TMPRSS5 | 205 | 0.100 | 0.1253 | Yes |
| 6 | CD44 | CD44 | 281 | 0.092 | 0.1383 | Yes |
| 7 | TIMP4 | TIMP4 | 300 | 0.090 | 0.1539 | Yes |
| 8 | GPRC5B | GPRC5B | 412 | 0.082 | 0.1635 | Yes |
| 9 | RGS20 | RGS20 | 422 | 0.081 | 0.1778 | Yes |
| 10 | PTPRZ1 | PTPRZ1 | 474 | 0.079 | 0.1897 | Yes |
| 11 | MAP2K5 | MAP2K5 | 598 | 0.073 | 0.1971 | Yes |
| 12 | RAP1GAP | RAP1GAP | 716 | 0.069 | 0.2041 | Yes |
| 13 | ABAT | ABAT | 806 | 0.066 | 0.2119 | Yes |
| 14 | FZD7 | FZD7 | 872 | 0.064 | 0.2203 | Yes |
| 15 | IDH1 | IDH1 | 946 | 0.062 | 0.2281 | Yes |
| 16 | STARD7 | STARD7 | 985 | 0.061 | 0.2374 | Yes |
| 17 | NLGN3 | NLGN3 | 1376 | 0.053 | 0.2288 | Yes |
| 18 | MARCKSL1 | MARCKSL1 | 1500 | 0.051 | 0.2323 | Yes |
| 19 | HIP1 | HIP1 | 1537 | 0.051 | 0.2398 | Yes |
| 20 | AKAP12 | AKAP12 | 1567 | 0.050 | 0.2475 | Yes |
| 21 | TRIM24 | TRIM24 | 1626 | 0.049 | 0.2537 | Yes |
| 22 | TTYH1 | TTYH1 | 1643 | 0.049 | 0.2619 | Yes |
| 23 | TBKBP1 | TBKBP1 | 2423 | 0.040 | 0.2328 | No |
| 24 | HOMER1 | HOMER1 | 2532 | 0.039 | 0.2348 | No |
| 25 | RASSF2 | RASSF2 | 2595 | 0.039 | 0.2389 | No |
| 26 | SLC38A3 | SLC38A3 | 2610 | 0.038 | 0.2452 | No |
| 27 | GAB1 | GAB1 | 2731 | 0.037 | 0.2464 | No |
| 28 | IRS1 | IRS1 | 2945 | 0.036 | 0.2429 | No |
| 29 | LGI2 | LGI2 | 3051 | 0.035 | 0.2444 | No |
| 30 | PIK3R2 | PIK3R2 | 3383 | 0.032 | 0.2348 | No |
| 31 | GPR19 | GPR19 | 3404 | 0.032 | 0.2397 | No |
| 32 | ART3 | ART3 | 3407 | 0.032 | 0.2454 | No |
| 33 | ANXA2P1 | ANXA2P1 | 3622 | 0.031 | 0.2410 | No |
| 34 | TUBA1A | TUBA1A | 3634 | 0.031 | 0.2460 | No |
| 35 | NME4 | NME4 | 3854 | 0.029 | 0.2410 | No |
| 36 | CSPG5 | CSPG5 | 4138 | 0.027 | 0.2328 | No |
| 37 | KANK1 | KANK1 | 4147 | 0.027 | 0.2374 | No |
| 38 | SLC4A4 | SLC4A4 | 4538 | 0.025 | 0.2237 | No |
| 39 | LMF1 | LMF1 | 4686 | 0.024 | 0.2212 | No |
| 40 | ELL2 | ELL2 | 4749 | 0.024 | 0.2226 | No |
| 41 | BMP2K | BMP2K | 4816 | 0.023 | 0.2238 | No |
| 42 | LAPTM4B | LAPTM4B | 4918 | 0.023 | 0.2232 | No |
| 43 | RLBP1 | RLBP1 | 4923 | 0.023 | 0.2272 | No |
| 44 | TUBB2B | TUBB2B | 5145 | 0.022 | 0.2208 | No |
| 45 | BCAN | BCAN | 5383 | 0.021 | 0.2135 | No |
| 46 | RBBP9 | RBBP9 | 5387 | 0.021 | 0.2170 | No |
| 47 | PNMA2 | PNMA2 | 5558 | 0.020 | 0.2127 | No |
| 48 | ATP9B | ATP9B | 5737 | 0.019 | 0.2078 | No |
| 49 | SOX11 | SOX11 | 5768 | 0.019 | 0.2098 | No |
| 50 | CCND2 | CCND2 | 5948 | 0.018 | 0.2047 | No |
| 51 | FAM114A1 | FAM114A1 | 6037 | 0.018 | 0.2038 | No |
| 52 | ATXN10 | ATXN10 | 6197 | 0.017 | 0.1994 | No |
| 53 | SRGN | SRGN | 6201 | 0.017 | 0.2023 | No |
| 54 | MMP16 | MMP16 | 6212 | 0.017 | 0.2049 | No |
| 55 | FSCN1 | FSCN1 | 6351 | 0.016 | 0.2014 | No |
| 56 | IFFO1 | IFFO1 | 6493 | 0.016 | 0.1977 | No |
| 57 | RAF1 | RAF1 | 7096 | 0.013 | 0.1720 | No |
| 58 | ENDOG | ENDOG | 7176 | 0.013 | 0.1706 | No |
| 59 | CREB5 | CREB5 | 7363 | 0.012 | 0.1641 | No |
| 60 | APBB2 | APBB2 | 7413 | 0.012 | 0.1640 | No |
| 61 | PHLPP1 | PHLPP1 | 7598 | 0.011 | 0.1574 | No |
| 62 | QDPR | QDPR | 7721 | 0.011 | 0.1536 | No |
| 63 | GLDC | GLDC | 8451 | 0.008 | 0.1211 | No |
| 64 | TRIB2 | TRIB2 | 8647 | 0.007 | 0.1133 | No |
| 65 | BCAR3 | BCAR3 | 8862 | 0.006 | 0.1045 | No |
| 66 | SYT11 | SYT11 | 8998 | 0.006 | 0.0993 | No |
| 67 | PODXL2 | PODXL2 | 9226 | 0.005 | 0.0896 | No |
| 68 | ST3GAL6 | ST3GAL6 | 9471 | 0.004 | 0.0790 | No |
| 69 | SLC19A3 | SLC19A3 | 9618 | 0.004 | 0.0729 | No |
| 70 | PRDM8 | PRDM8 | 10100 | 0.002 | 0.0508 | No |
| 71 | SDC2 | SDC2 | 10134 | 0.002 | 0.0496 | No |
| 72 | NKX2-2 | NKX2-2 | 10172 | 0.002 | 0.0482 | No |
| 73 | ASCL1 | ASCL1 | 10271 | 0.001 | 0.0439 | No |
| 74 | SPATA6 | SPATA6 | 10570 | 0.000 | 0.0301 | No |
| 75 | NFAT5 | NFAT5 | 10594 | 0.000 | 0.0290 | No |
| 76 | HLA-DPA1 | HLA-DPA1 | 10851 | -0.001 | 0.0172 | No |
| 77 | GPD1L | GPD1L | 11158 | -0.002 | 0.0033 | No |
| 78 | RBM38 | RBM38 | 11197 | -0.002 | 0.0018 | No |
| 79 | TRAF4 | TRAF4 | 11386 | -0.003 | -0.0065 | No |
| 80 | VEGFC | VEGFC | 11392 | -0.003 | -0.0062 | No |
| 81 | MYOF | MYOF | 11422 | -0.003 | -0.0071 | No |
| 82 | NOVA1 | NOVA1 | 11602 | -0.003 | -0.0149 | No |
| 83 | C1orf61 | C1orf61 | 12173 | -0.005 | -0.0405 | No |
| 84 | POLR2F | POLR2F | 12347 | -0.006 | -0.0475 | No |
| 85 | PPFIBP2 | PPFIBP2 | 12472 | -0.006 | -0.0521 | No |
| 86 | GCNT1 | GCNT1 | 12523 | -0.007 | -0.0533 | No |
| 87 | RAB27A | RAB27A | 12626 | -0.007 | -0.0568 | No |
| 88 | PTGS2 | PTGS2 | 13295 | -0.009 | -0.0862 | No |
| 89 | COL4A6 | COL4A6 | 13642 | -0.011 | -0.1004 | No |
| 90 | DUSP6 | DUSP6 | 13939 | -0.012 | -0.1121 | No |
| 91 | MFAP2 | MFAP2 | 14033 | -0.012 | -0.1142 | No |
| 92 | VASH1 | VASH1 | 14254 | -0.013 | -0.1221 | No |
| 93 | BNC2 | BNC2 | 14603 | -0.014 | -0.1358 | No |
| 94 | SLIT2 | SLIT2 | 14812 | -0.015 | -0.1427 | No |
| 95 | ZNF544 | ZNF544 | 15182 | -0.017 | -0.1569 | No |
| 96 | NES | NES | 15616 | -0.019 | -0.1737 | No |
| 97 | PTGER4 | PTGER4 | 15657 | -0.019 | -0.1721 | No |
| 98 | TBC1D22B | TBC1D22B | 15797 | -0.020 | -0.1751 | No |
| 99 | FRMPD1 | FRMPD1 | 16019 | -0.021 | -0.1817 | No |
| 100 | S100B | S100B | 16586 | -0.023 | -0.2039 | No |
| 101 | ATP6V0E2 | ATP6V0E2 | 17041 | -0.025 | -0.2204 | No |
| 102 | RASL12 | RASL12 | 17174 | -0.026 | -0.2218 | No |
| 103 | CHST11 | CHST11 | 17182 | -0.026 | -0.2174 | No |
| 104 | SYT17 | SYT17 | 17215 | -0.027 | -0.2141 | No |
| 105 | BAALC | BAALC | 17290 | -0.027 | -0.2127 | No |
| 106 | GPR68 | GPR68 | 17317 | -0.027 | -0.2090 | No |
| 107 | CGREF1 | CGREF1 | 17387 | -0.027 | -0.2073 | No |
| 108 | BCHE | BCHE | 17823 | -0.030 | -0.2222 | No |
| 109 | GNAZ | GNAZ | 17833 | -0.030 | -0.2172 | No |
| 110 | RRAS2 | RRAS2 | 17842 | -0.030 | -0.2121 | No |
| 111 | SNX27 | SNX27 | 17859 | -0.030 | -0.2075 | No |
| 112 | DBI | DBI | 18135 | -0.032 | -0.2145 | No |
| 113 | RAPGEF4 | RAPGEF4 | 18137 | -0.032 | -0.2088 | No |
| 114 | NINJ2 | NINJ2 | 18188 | -0.032 | -0.2053 | No |
| 115 | MAP3K1 | MAP3K1 | 18630 | -0.035 | -0.2195 | No |
| 116 | VGLL4 | VGLL4 | 18637 | -0.036 | -0.2133 | No |
| 117 | PLLP | PLLP | 18644 | -0.036 | -0.2072 | No |
| 118 | LEPROTL1 | LEPROTL1 | 18666 | -0.036 | -0.2017 | No |
| 119 | CCDC28B | CCDC28B | 18891 | -0.038 | -0.2053 | No |
| 120 | GOLM1 | GOLM1 | 18963 | -0.038 | -0.2017 | No |
| 121 | TMEM206 | TMEM206 | 19314 | -0.042 | -0.2105 | No |
| 122 | P4HA2 | P4HA2 | 19472 | -0.043 | -0.2100 | No |
| 123 | CAST | CAST | 19519 | -0.044 | -0.2042 | No |
| 124 | SLC10A3 | SLC10A3 | 19691 | -0.046 | -0.2039 | No |
| 125 | DNAH9 | DNAH9 | 19721 | -0.046 | -0.1969 | No |
| 126 | CSPG4 | CSPG4 | 19771 | -0.047 | -0.1908 | No |
| 127 | FAM131B | FAM131B | 19876 | -0.048 | -0.1869 | No |
| 128 | HES1 | HES1 | 19987 | -0.050 | -0.1831 | No |
| 129 | CITED1 | CITED1 | 20119 | -0.051 | -0.1799 | No |
| 130 | TMEM121 | TMEM121 | 20358 | -0.056 | -0.1809 | No |
| 131 | ETV5 | ETV5 | 20396 | -0.056 | -0.1725 | No |
| 132 | GAS1 | GAS1 | 20486 | -0.058 | -0.1662 | No |
| 133 | CYP1B1 | CYP1B1 | 20518 | -0.059 | -0.1570 | No |
| 134 | ZYX | ZYX | 20520 | -0.059 | -0.1465 | No |
| 135 | MYO1C | MYO1C | 20807 | -0.066 | -0.1478 | No |
| 136 | GNG7 | GNG7 | 20829 | -0.067 | -0.1366 | No |
| 137 | SHC3 | SHC3 | 20860 | -0.068 | -0.1257 | No |
| 138 | PHACTR2 | PHACTR2 | 21040 | -0.074 | -0.1206 | No |
| 139 | ARC | ARC | 21434 | -0.109 | -0.1192 | No |
| 140 | IL1A | IL1A | 21453 | -0.113 | -0.0996 | No |
| 141 | IRF1 | IRF1 | 21459 | -0.115 | -0.0790 | No |
| 142 | PICK1 | PICK1 | 21467 | -0.117 | -0.0582 | No |
| 143 | NDE1 | NDE1 | 21509 | -0.142 | -0.0345 | No |
| 144 | COL7A1 | COL7A1 | 21579 | -0.213 | 0.0007 | No |