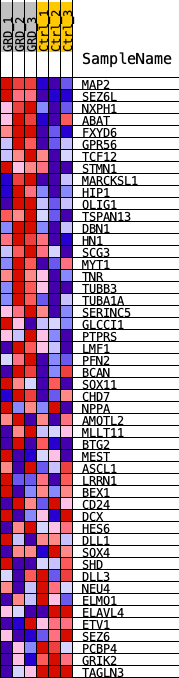
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | GRD |
| GeneSet | NEFTEL_NPC1_2019 |
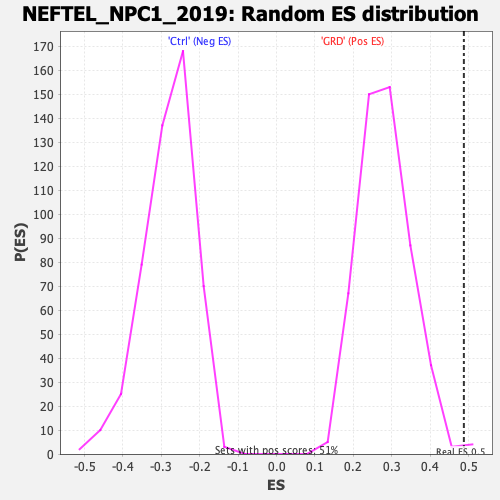
| Enrichment Score (ES) | 0.48781687 |
| Normalized Enrichment Score (NES) | 1.7288727 |
| Nominal p-value | 0.007905139 |
| FDR q-value | 0.0046717934 |
| FWER p-Value | 0.021 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MAP2 | MAP2 | 145 | 0.108 | 0.0576 | Yes |
| 2 | SEZ6L | SEZ6L | 228 | 0.096 | 0.1110 | Yes |
| 3 | NXPH1 | NXPH1 | 253 | 0.094 | 0.1655 | Yes |
| 4 | ABAT | ABAT | 806 | 0.066 | 0.1788 | Yes |
| 5 | FXYD6 | FXYD6 | 892 | 0.063 | 0.2123 | Yes |
| 6 | GPR56 | GPR56 | 1019 | 0.060 | 0.2422 | Yes |
| 7 | TCF12 | TCF12 | 1195 | 0.057 | 0.2677 | Yes |
| 8 | STMN1 | STMN1 | 1330 | 0.054 | 0.2935 | Yes |
| 9 | MARCKSL1 | MARCKSL1 | 1500 | 0.051 | 0.3160 | Yes |
| 10 | HIP1 | HIP1 | 1537 | 0.051 | 0.3444 | Yes |
| 11 | OLIG1 | OLIG1 | 1582 | 0.050 | 0.3722 | Yes |
| 12 | TSPAN13 | TSPAN13 | 1696 | 0.049 | 0.3958 | Yes |
| 13 | DBN1 | DBN1 | 1852 | 0.046 | 0.4161 | Yes |
| 14 | HN1 | HN1 | 1966 | 0.045 | 0.4377 | Yes |
| 15 | SCG3 | SCG3 | 2487 | 0.039 | 0.4369 | Yes |
| 16 | MYT1 | MYT1 | 3053 | 0.035 | 0.4315 | Yes |
| 17 | TNR | TNR | 3179 | 0.034 | 0.4457 | Yes |
| 18 | TUBB3 | TUBB3 | 3623 | 0.031 | 0.4434 | Yes |
| 19 | TUBA1A | TUBA1A | 3634 | 0.031 | 0.4611 | Yes |
| 20 | SERINC5 | SERINC5 | 3806 | 0.029 | 0.4706 | Yes |
| 21 | GLCCI1 | GLCCI1 | 4470 | 0.025 | 0.4550 | Yes |
| 22 | PTPRS | PTPRS | 4634 | 0.024 | 0.4619 | Yes |
| 23 | LMF1 | LMF1 | 4686 | 0.024 | 0.4739 | Yes |
| 24 | PFN2 | PFN2 | 4694 | 0.024 | 0.4878 | Yes |
| 25 | BCAN | BCAN | 5383 | 0.021 | 0.4681 | No |
| 26 | SOX11 | SOX11 | 5768 | 0.019 | 0.4615 | No |
| 27 | CHD7 | CHD7 | 6015 | 0.018 | 0.4605 | No |
| 28 | NPPA | NPPA | 7771 | 0.011 | 0.3854 | No |
| 29 | AMOTL2 | AMOTL2 | 7900 | 0.010 | 0.3854 | No |
| 30 | MLLT11 | MLLT11 | 8092 | 0.009 | 0.3820 | No |
| 31 | BTG2 | BTG2 | 8789 | 0.007 | 0.3536 | No |
| 32 | MEST | MEST | 9440 | 0.004 | 0.3261 | No |
| 33 | ASCL1 | ASCL1 | 10271 | 0.001 | 0.2883 | No |
| 34 | LRRN1 | LRRN1 | 11449 | -0.003 | 0.2354 | No |
| 35 | BEX1 | BEX1 | 12138 | -0.005 | 0.2065 | No |
| 36 | CD24 | CD24 | 12357 | -0.006 | 0.1999 | No |
| 37 | DCX | DCX | 12388 | -0.006 | 0.2021 | No |
| 38 | HES6 | HES6 | 12547 | -0.007 | 0.1986 | No |
| 39 | DLL1 | DLL1 | 12760 | -0.007 | 0.1932 | No |
| 40 | SOX4 | SOX4 | 13444 | -0.010 | 0.1673 | No |
| 41 | SHD | SHD | 14332 | -0.013 | 0.1341 | No |
| 42 | DLL3 | DLL3 | 14605 | -0.014 | 0.1300 | No |
| 43 | NEU4 | NEU4 | 16227 | -0.022 | 0.0675 | No |
| 44 | ELMO1 | ELMO1 | 17786 | -0.030 | 0.0128 | No |
| 45 | ELAVL4 | ELAVL4 | 17794 | -0.030 | 0.0300 | No |
| 46 | ETV1 | ETV1 | 18514 | -0.035 | 0.0172 | No |
| 47 | SEZ6 | SEZ6 | 18974 | -0.038 | 0.0188 | No |
| 48 | PCBP4 | PCBP4 | 20027 | -0.050 | -0.0002 | No |
| 49 | GRIK2 | GRIK2 | 20588 | -0.060 | 0.0096 | No |
| 50 | TAGLN3 | TAGLN3 | 20667 | -0.062 | 0.0431 | No |