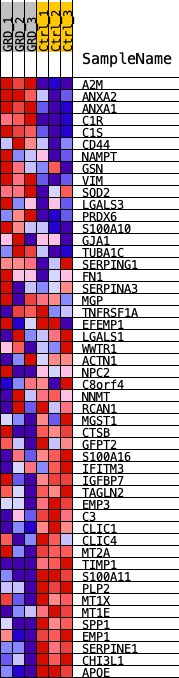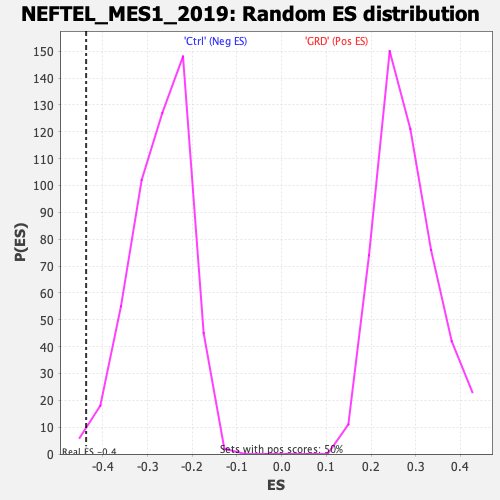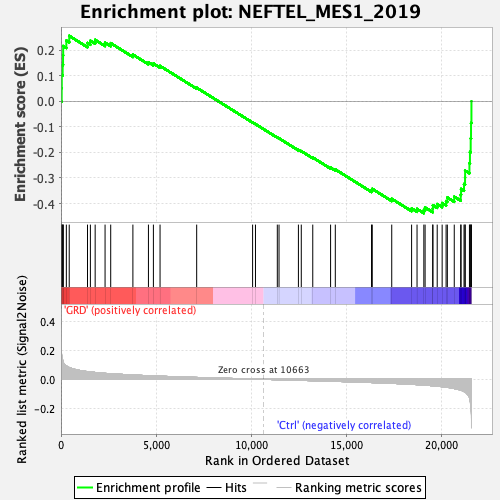
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | NEFTEL_MES1_2019 |
| Enrichment Score (ES) | -0.4376462 |
| Normalized Enrichment Score (NES) | -1.6074172 |
| Nominal p-value | 0.009940358 |
| FDR q-value | 0.00918006 |
| FWER p-Value | 0.071 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | A2M | A2M | 36 | 0.168 | 0.0513 | No |
| 2 | ANXA2 | ANXA2 | 41 | 0.164 | 0.1028 | No |
| 3 | ANXA1 | ANXA1 | 87 | 0.133 | 0.1428 | No |
| 4 | C1R | C1R | 108 | 0.120 | 0.1798 | No |
| 5 | C1S | C1S | 115 | 0.117 | 0.2164 | No |
| 6 | CD44 | CD44 | 281 | 0.092 | 0.2377 | No |
| 7 | NAMPT | NAMPT | 433 | 0.081 | 0.2562 | No |
| 8 | GSN | GSN | 1394 | 0.053 | 0.2283 | No |
| 9 | VIM | VIM | 1546 | 0.051 | 0.2372 | No |
| 10 | SOD2 | SOD2 | 1797 | 0.047 | 0.2405 | No |
| 11 | LGALS3 | LGALS3 | 2318 | 0.041 | 0.2293 | No |
| 12 | PRDX6 | PRDX6 | 2615 | 0.038 | 0.2277 | No |
| 13 | S100A10 | S100A10 | 3782 | 0.030 | 0.1829 | No |
| 14 | GJA1 | GJA1 | 4600 | 0.025 | 0.1527 | No |
| 15 | TUBA1C | TUBA1C | 4861 | 0.023 | 0.1480 | No |
| 16 | SERPING1 | SERPING1 | 5211 | 0.021 | 0.1386 | No |
| 17 | FN1 | FN1 | 7138 | 0.013 | 0.0532 | No |
| 18 | SERPINA3 | SERPINA3 | 10080 | 0.002 | -0.0826 | No |
| 19 | MGP | MGP | 10231 | 0.002 | -0.0891 | No |
| 20 | TNFRSF1A | TNFRSF1A | 11380 | -0.003 | -0.1416 | No |
| 21 | EFEMP1 | EFEMP1 | 11470 | -0.003 | -0.1448 | No |
| 22 | LGALS1 | LGALS1 | 12490 | -0.006 | -0.1901 | No |
| 23 | WWTR1 | WWTR1 | 12641 | -0.007 | -0.1949 | No |
| 24 | ACTN1 | ACTN1 | 13245 | -0.009 | -0.2200 | No |
| 25 | NPC2 | NPC2 | 14183 | -0.013 | -0.2594 | No |
| 26 | C8orf4 | C8orf4 | 14431 | -0.014 | -0.2666 | No |
| 27 | NNMT | NNMT | 16336 | -0.022 | -0.3480 | No |
| 28 | RCAN1 | RCAN1 | 16375 | -0.022 | -0.3428 | No |
| 29 | MGST1 | MGST1 | 17400 | -0.027 | -0.3816 | No |
| 30 | CTSB | CTSB | 18445 | -0.034 | -0.4193 | No |
| 31 | GFPT2 | GFPT2 | 18731 | -0.036 | -0.4211 | No |
| 32 | S100A16 | S100A16 | 19089 | -0.039 | -0.4252 | Yes |
| 33 | IFITM3 | IFITM3 | 19155 | -0.040 | -0.4155 | Yes |
| 34 | IGFBP7 | IGFBP7 | 19557 | -0.044 | -0.4202 | Yes |
| 35 | TAGLN2 | TAGLN2 | 19565 | -0.044 | -0.4065 | Yes |
| 36 | EMP3 | EMP3 | 19792 | -0.047 | -0.4022 | Yes |
| 37 | C3 | C3 | 20052 | -0.051 | -0.3982 | Yes |
| 38 | CLIC1 | CLIC1 | 20260 | -0.054 | -0.3908 | Yes |
| 39 | CLIC4 | CLIC4 | 20326 | -0.055 | -0.3764 | Yes |
| 40 | MT2A | MT2A | 20689 | -0.063 | -0.3732 | Yes |
| 41 | TIMP1 | TIMP1 | 21029 | -0.074 | -0.3657 | Yes |
| 42 | S100A11 | S100A11 | 21047 | -0.075 | -0.3429 | Yes |
| 43 | PLP2 | PLP2 | 21203 | -0.083 | -0.3238 | Yes |
| 44 | MT1X | MT1X | 21257 | -0.087 | -0.2987 | Yes |
| 45 | MT1E | MT1E | 21261 | -0.087 | -0.2712 | Yes |
| 46 | SPP1 | SPP1 | 21488 | -0.124 | -0.2424 | Yes |
| 47 | EMP1 | EMP1 | 21515 | -0.145 | -0.1980 | Yes |
| 48 | SERPINE1 | SERPINE1 | 21554 | -0.173 | -0.1451 | Yes |
| 49 | CHI3L1 | CHI3L1 | 21572 | -0.194 | -0.0847 | Yes |
| 50 | APOE | APOE | 21593 | -0.272 | 0.0001 | Yes |