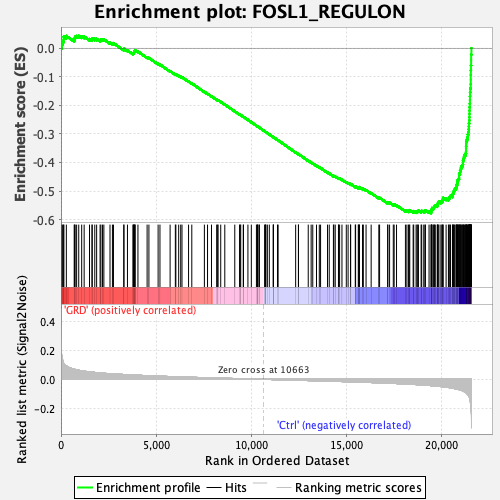
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | FOSL1_REGULON |
| Enrichment Score (ES) | -0.57744354 |
| Normalized Enrichment Score (NES) | -2.627471 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ANXA2 | ANXA2 | 41 | 0.164 | 0.0131 | No |
| 2 | ANXA1 | ANXA1 | 87 | 0.133 | 0.0233 | No |
| 3 | CTNND2 | CTNND2 | 142 | 0.109 | 0.0308 | No |
| 4 | MAP2 | MAP2 | 145 | 0.108 | 0.0407 | No |
| 5 | CD44 | CD44 | 281 | 0.092 | 0.0428 | No |
| 6 | UPP1 | UPP1 | 694 | 0.070 | 0.0300 | No |
| 7 | ANXA2P3 | ANXA2P3 | 715 | 0.069 | 0.0354 | No |
| 8 | PARVA | PARVA | 744 | 0.068 | 0.0403 | No |
| 9 | VCL | VCL | 822 | 0.065 | 0.0427 | No |
| 10 | FLNA | FLNA | 933 | 0.062 | 0.0433 | No |
| 11 | CDKN1B | CDKN1B | 1088 | 0.059 | 0.0415 | No |
| 12 | TRAM2 | TRAM2 | 1218 | 0.056 | 0.0406 | No |
| 13 | MARCKSL1 | MARCKSL1 | 1500 | 0.051 | 0.0322 | No |
| 14 | ANXA2P2 | ANXA2P2 | 1630 | 0.049 | 0.0307 | No |
| 15 | PQLC3 | PQLC3 | 1642 | 0.049 | 0.0347 | No |
| 16 | IMPAD1 | IMPAD1 | 1777 | 0.047 | 0.0328 | No |
| 17 | ZMYM4 | ZMYM4 | 1875 | 0.046 | 0.0325 | No |
| 18 | ZBTB38 | ZBTB38 | 2056 | 0.044 | 0.0281 | No |
| 19 | SHC1 | SHC1 | 2099 | 0.044 | 0.0302 | No |
| 20 | CD46 | CD46 | 2185 | 0.043 | 0.0301 | No |
| 21 | DEGS1 | DEGS1 | 2255 | 0.042 | 0.0307 | No |
| 22 | MAST1 | MAST1 | 2578 | 0.039 | 0.0193 | No |
| 23 | ANPEP | ANPEP | 2706 | 0.038 | 0.0168 | No |
| 24 | GBE1 | GBE1 | 2760 | 0.037 | 0.0177 | No |
| 25 | STC1 | STC1 | 3303 | 0.033 | -0.0046 | No |
| 26 | PLCB1 | PLCB1 | 3312 | 0.033 | -0.0019 | No |
| 27 | BET1L | BET1L | 3496 | 0.032 | -0.0076 | No |
| 28 | S100A10 | S100A10 | 3782 | 0.030 | -0.0182 | No |
| 29 | TPM2 | TPM2 | 3817 | 0.029 | -0.0171 | No |
| 30 | MAFG | MAFG | 3836 | 0.029 | -0.0153 | No |
| 31 | AKR1C3 | AKR1C3 | 3844 | 0.029 | -0.0129 | No |
| 32 | ABCD1 | ABCD1 | 3858 | 0.029 | -0.0108 | No |
| 33 | MYD88 | MYD88 | 3892 | 0.029 | -0.0097 | No |
| 34 | BACE2 | BACE2 | 3897 | 0.029 | -0.0073 | No |
| 35 | EXOC3 | EXOC3 | 4048 | 0.028 | -0.0117 | No |
| 36 | TCF4 | TCF4 | 4530 | 0.025 | -0.0319 | No |
| 37 | IL13RA1 | IL13RA1 | 4625 | 0.025 | -0.0340 | No |
| 38 | CLIP4 | CLIP4 | 5109 | 0.022 | -0.0546 | No |
| 39 | CRIM1 | CRIM1 | 5206 | 0.022 | -0.0571 | No |
| 40 | WDR1 | WDR1 | 5740 | 0.019 | -0.0803 | No |
| 41 | CHD7 | CHD7 | 6015 | 0.018 | -0.0915 | No |
| 42 | ADRM1 | ADRM1 | 6043 | 0.018 | -0.0912 | No |
| 43 | MAN1A1 | MAN1A1 | 6191 | 0.017 | -0.0965 | No |
| 44 | FUT9 | FUT9 | 6298 | 0.016 | -0.0999 | No |
| 45 | B3GNT2 | B3GNT2 | 6365 | 0.016 | -0.1015 | No |
| 46 | CAV2 | CAV2 | 6711 | 0.015 | -0.1163 | No |
| 47 | SMAGP | SMAGP | 6881 | 0.014 | -0.1229 | No |
| 48 | AXL | AXL | 7543 | 0.011 | -0.1528 | No |
| 49 | LRRFIP2 | LRRFIP2 | 7709 | 0.011 | -0.1595 | No |
| 50 | DSTN | DSTN | 7917 | 0.010 | -0.1683 | No |
| 51 | BFSP1 | BFSP1 | 8190 | 0.009 | -0.1802 | No |
| 52 | SLC38A10 | SLC38A10 | 8246 | 0.009 | -0.1820 | No |
| 53 | GLIPR1 | GLIPR1 | 8258 | 0.009 | -0.1817 | No |
| 54 | TPD52L2 | TPD52L2 | 8403 | 0.008 | -0.1877 | No |
| 55 | GLT8D2 | GLT8D2 | 8617 | 0.007 | -0.1970 | No |
| 56 | BAG3 | BAG3 | 9142 | 0.005 | -0.2210 | No |
| 57 | MCFD2 | MCFD2 | 9394 | 0.005 | -0.2323 | No |
| 58 | HSPA5 | HSPA5 | 9422 | 0.004 | -0.2332 | No |
| 59 | RAC2 | RAC2 | 9455 | 0.004 | -0.2343 | No |
| 60 | CYB561 | CYB561 | 9589 | 0.004 | -0.2401 | No |
| 61 | ANXA11 | ANXA11 | 9600 | 0.004 | -0.2403 | No |
| 62 | RHOG | RHOG | 9834 | 0.003 | -0.2509 | No |
| 63 | TMEM109 | TMEM109 | 10013 | 0.002 | -0.2590 | No |
| 64 | SEMA4F | SEMA4F | 10281 | 0.001 | -0.2714 | No |
| 65 | PMAIP1 | PMAIP1 | 10328 | 0.001 | -0.2734 | No |
| 66 | KCNN4 | KCNN4 | 10334 | 0.001 | -0.2736 | No |
| 67 | ARFRP1 | ARFRP1 | 10353 | 0.001 | -0.2743 | No |
| 68 | COL6A2 | COL6A2 | 10417 | 0.001 | -0.2772 | No |
| 69 | LGALS8 | LGALS8 | 10438 | 0.001 | -0.2780 | No |
| 70 | SERINC3 | SERINC3 | 10713 | -0.000 | -0.2908 | No |
| 71 | PRNP | PRNP | 10742 | -0.000 | -0.2921 | No |
| 72 | KCND2 | KCND2 | 10763 | -0.000 | -0.2930 | No |
| 73 | MGAT4B | MGAT4B | 10778 | -0.000 | -0.2937 | No |
| 74 | TNFRSF10B | TNFRSF10B | 10856 | -0.001 | -0.2972 | No |
| 75 | RRBP1 | RRBP1 | 10959 | -0.001 | -0.3019 | No |
| 76 | OLFML2B | OLFML2B | 11166 | -0.002 | -0.3114 | No |
| 77 | ESYT1 | ESYT1 | 11172 | -0.002 | -0.3114 | No |
| 78 | FOSL2 | FOSL2 | 11178 | -0.002 | -0.3115 | No |
| 79 | APOBEC3B | APOBEC3B | 11389 | -0.003 | -0.3211 | No |
| 80 | MYOF | MYOF | 11422 | -0.003 | -0.3223 | No |
| 81 | S100A13 | S100A13 | 12345 | -0.006 | -0.3649 | No |
| 82 | LGALS1 | LGALS1 | 12490 | -0.006 | -0.3711 | No |
| 83 | CXXC4 | CXXC4 | 12495 | -0.006 | -0.3707 | No |
| 84 | FOXC1 | FOXC1 | 13007 | -0.008 | -0.3938 | No |
| 85 | MET | MET | 13160 | -0.009 | -0.4001 | No |
| 86 | ACTN1 | ACTN1 | 13245 | -0.009 | -0.4032 | No |
| 87 | RAVER2 | RAVER2 | 13442 | -0.010 | -0.4115 | No |
| 88 | FLII | FLII | 13589 | -0.010 | -0.4173 | No |
| 89 | ITGA6 | ITGA6 | 13656 | -0.011 | -0.4194 | No |
| 90 | RELB | RELB | 14022 | -0.012 | -0.4354 | No |
| 91 | LPP | LPP | 14112 | -0.013 | -0.4384 | No |
| 92 | FBRS | FBRS | 14331 | -0.013 | -0.4474 | No |
| 93 | KLHL21 | KLHL21 | 14354 | -0.013 | -0.4472 | No |
| 94 | CAV1 | CAV1 | 14427 | -0.014 | -0.4493 | No |
| 95 | ANXA4 | ANXA4 | 14597 | -0.014 | -0.4559 | No |
| 96 | RNH1 | RNH1 | 14608 | -0.014 | -0.4550 | No |
| 97 | ANP32A | ANP32A | 14657 | -0.015 | -0.4559 | No |
| 98 | IL18R1 | IL18R1 | 14781 | -0.015 | -0.4603 | No |
| 99 | TXN | TXN | 15009 | -0.016 | -0.4694 | No |
| 100 | FADS3 | FADS3 | 15104 | -0.017 | -0.4723 | No |
| 101 | ACTN4 | ACTN4 | 15234 | -0.017 | -0.4767 | No |
| 102 | RPS6KA4 | RPS6KA4 | 15237 | -0.017 | -0.4753 | No |
| 103 | ARSA | ARSA | 15481 | -0.018 | -0.4850 | No |
| 104 | VDR | VDR | 15499 | -0.018 | -0.4841 | No |
| 105 | PIGT | PIGT | 15640 | -0.019 | -0.4889 | No |
| 106 | PTGER4 | PTGER4 | 15657 | -0.019 | -0.4879 | No |
| 107 | GEM | GEM | 15692 | -0.019 | -0.4878 | No |
| 108 | FOXN3 | FOXN3 | 15699 | -0.019 | -0.4863 | No |
| 109 | MYH9 | MYH9 | 15863 | -0.020 | -0.4921 | No |
| 110 | FHL2 | FHL2 | 15905 | -0.020 | -0.4922 | No |
| 111 | KCND1 | KCND1 | 16049 | -0.021 | -0.4970 | No |
| 112 | VASP | VASP | 16317 | -0.022 | -0.5074 | No |
| 113 | SRPX2 | SRPX2 | 16721 | -0.024 | -0.5241 | No |
| 114 | IFRD2 | IFRD2 | 16761 | -0.024 | -0.5237 | No |
| 115 | GSTA4 | GSTA4 | 17184 | -0.026 | -0.5410 | No |
| 116 | EDEM1 | EDEM1 | 17197 | -0.026 | -0.5391 | No |
| 117 | REXO2 | REXO2 | 17287 | -0.027 | -0.5408 | No |
| 118 | TAPBP | TAPBP | 17481 | -0.028 | -0.5473 | No |
| 119 | PERP | PERP | 17526 | -0.028 | -0.5467 | No |
| 120 | MGAT1 | MGAT1 | 17652 | -0.029 | -0.5499 | No |
| 121 | TMED9 | TMED9 | 18120 | -0.032 | -0.5689 | No |
| 122 | SERPINB6 | SERPINB6 | 18196 | -0.032 | -0.5694 | No |
| 123 | CBX7 | CBX7 | 18278 | -0.033 | -0.5702 | No |
| 124 | MVP | MVP | 18318 | -0.033 | -0.5689 | No |
| 125 | YIPF3 | YIPF3 | 18348 | -0.033 | -0.5672 | No |
| 126 | MYLK | MYLK | 18526 | -0.035 | -0.5723 | No |
| 127 | DNAJC10 | DNAJC10 | 18556 | -0.035 | -0.5705 | No |
| 128 | CSRP1 | CSRP1 | 18681 | -0.036 | -0.5730 | No |
| 129 | GFPT2 | GFPT2 | 18731 | -0.036 | -0.5719 | No |
| 130 | SSH3 | SSH3 | 18771 | -0.037 | -0.5704 | No |
| 131 | SH3BGRL3 | SH3BGRL3 | 18804 | -0.037 | -0.5685 | No |
| 132 | TSPAN7 | TSPAN7 | 18945 | -0.038 | -0.5715 | No |
| 133 | KCNAB2 | KCNAB2 | 18977 | -0.038 | -0.5694 | No |
| 134 | LMNA | LMNA | 19085 | -0.039 | -0.5708 | No |
| 135 | ATP1A1 | ATP1A1 | 19146 | -0.040 | -0.5699 | No |
| 136 | KDELR3 | KDELR3 | 19172 | -0.040 | -0.5674 | No |
| 137 | GSDMD | GSDMD | 19363 | -0.042 | -0.5724 | No |
| 138 | P4HA2 | P4HA2 | 19472 | -0.043 | -0.5735 | Yes |
| 139 | WDR55 | WDR55 | 19475 | -0.043 | -0.5696 | Yes |
| 140 | CRTAM | CRTAM | 19489 | -0.043 | -0.5662 | Yes |
| 141 | CAPN2 | CAPN2 | 19513 | -0.044 | -0.5632 | Yes |
| 142 | CAST | CAST | 19519 | -0.044 | -0.5595 | Yes |
| 143 | PLAUR | PLAUR | 19606 | -0.045 | -0.5594 | Yes |
| 144 | FADD | FADD | 19627 | -0.045 | -0.5561 | Yes |
| 145 | CDC42EP3 | CDC42EP3 | 19644 | -0.045 | -0.5527 | Yes |
| 146 | SLC10A3 | SLC10A3 | 19691 | -0.046 | -0.5507 | Yes |
| 147 | EMP3 | EMP3 | 19792 | -0.047 | -0.5510 | Yes |
| 148 | TMEM214 | TMEM214 | 19809 | -0.047 | -0.5474 | Yes |
| 149 | QSOX1 | QSOX1 | 19823 | -0.047 | -0.5437 | Yes |
| 150 | PARP12 | PARP12 | 19840 | -0.048 | -0.5400 | Yes |
| 151 | MOCOS | MOCOS | 19883 | -0.048 | -0.5375 | Yes |
| 152 | PXN | PXN | 19986 | -0.050 | -0.5378 | Yes |
| 153 | PARVB | PARVB | 20036 | -0.050 | -0.5354 | Yes |
| 154 | FYCO1 | FYCO1 | 20080 | -0.051 | -0.5327 | Yes |
| 155 | DLX2 | DLX2 | 20090 | -0.051 | -0.5284 | Yes |
| 156 | ITGA5 | ITGA5 | 20099 | -0.051 | -0.5241 | Yes |
| 157 | CLIC1 | CLIC1 | 20260 | -0.054 | -0.5266 | Yes |
| 158 | ETHE1 | ETHE1 | 20382 | -0.056 | -0.5271 | Yes |
| 159 | ADSS | ADSS | 20415 | -0.057 | -0.5234 | Yes |
| 160 | CUL7 | CUL7 | 20458 | -0.057 | -0.5201 | Yes |
| 161 | CYBRD1 | CYBRD1 | 20489 | -0.058 | -0.5162 | Yes |
| 162 | NSMAF | NSMAF | 20593 | -0.060 | -0.5155 | Yes |
| 163 | PLEKHF1 | PLEKHF1 | 20606 | -0.061 | -0.5104 | Yes |
| 164 | TFPI2 | TFPI2 | 20632 | -0.062 | -0.5059 | Yes |
| 165 | SQSTM1 | SQSTM1 | 20653 | -0.062 | -0.5012 | Yes |
| 166 | MT2A | MT2A | 20689 | -0.063 | -0.4970 | Yes |
| 167 | RBKS | RBKS | 20720 | -0.064 | -0.4925 | Yes |
| 168 | COQ10B | COQ10B | 20781 | -0.066 | -0.4893 | Yes |
| 169 | MYO1C | MYO1C | 20807 | -0.066 | -0.4843 | Yes |
| 170 | RNFT2 | RNFT2 | 20811 | -0.067 | -0.4783 | Yes |
| 171 | GLRX | GLRX | 20844 | -0.068 | -0.4736 | Yes |
| 172 | S100A6 | S100A6 | 20850 | -0.068 | -0.4676 | Yes |
| 173 | SH2D4A | SH2D4A | 20863 | -0.068 | -0.4619 | Yes |
| 174 | AHNAK | AHNAK | 20906 | -0.070 | -0.4574 | Yes |
| 175 | PNPLA2 | PNPLA2 | 20942 | -0.071 | -0.4526 | Yes |
| 176 | RIN1 | RIN1 | 20944 | -0.071 | -0.4461 | Yes |
| 177 | YIF1A | YIF1A | 20945 | -0.071 | -0.4396 | Yes |
| 178 | SLC35C1 | SLC35C1 | 20992 | -0.072 | -0.4350 | Yes |
| 179 | ADORA2B | ADORA2B | 20998 | -0.073 | -0.4286 | Yes |
| 180 | TIMP1 | TIMP1 | 21029 | -0.074 | -0.4232 | Yes |
| 181 | S100A11 | S100A11 | 21047 | -0.075 | -0.4171 | Yes |
| 182 | DRAP1 | DRAP1 | 21080 | -0.076 | -0.4116 | Yes |
| 183 | IQGAP1 | IQGAP1 | 21134 | -0.079 | -0.4068 | Yes |
| 184 | TSPAN1 | TSPAN1 | 21147 | -0.080 | -0.4000 | Yes |
| 185 | PHLDA2 | PHLDA2 | 21151 | -0.080 | -0.3928 | Yes |
| 186 | ARPC1B | ARPC1B | 21177 | -0.082 | -0.3865 | Yes |
| 187 | PLP2 | PLP2 | 21203 | -0.083 | -0.3800 | Yes |
| 188 | DRAM1 | DRAM1 | 21232 | -0.086 | -0.3734 | Yes |
| 189 | CEBPB | CEBPB | 21289 | -0.090 | -0.3677 | Yes |
| 190 | MICAL2 | MICAL2 | 21308 | -0.091 | -0.3601 | Yes |
| 191 | FURIN | FURIN | 21311 | -0.092 | -0.3518 | Yes |
| 192 | EPHA2 | EPHA2 | 21313 | -0.092 | -0.3434 | Yes |
| 193 | SH2B3 | SH2B3 | 21319 | -0.093 | -0.3351 | Yes |
| 194 | PROCR | PROCR | 21321 | -0.093 | -0.3265 | Yes |
| 195 | SYNJ2 | SYNJ2 | 21354 | -0.098 | -0.3190 | Yes |
| 196 | DCBLD2 | DCBLD2 | 21387 | -0.103 | -0.3111 | Yes |
| 197 | LIF | LIF | 21404 | -0.105 | -0.3021 | Yes |
| 198 | WIPI1 | WIPI1 | 21433 | -0.109 | -0.2934 | Yes |
| 199 | NT5E | NT5E | 21448 | -0.112 | -0.2838 | Yes |
| 200 | G6PD | G6PD | 21456 | -0.114 | -0.2736 | Yes |
| 201 | PRSS23 | PRSS23 | 21462 | -0.116 | -0.2631 | Yes |
| 202 | IER3 | IER3 | 21480 | -0.122 | -0.2527 | Yes |
| 203 | COL18A1 | COL18A1 | 21486 | -0.124 | -0.2415 | Yes |
| 204 | SDC4 | SDC4 | 21489 | -0.125 | -0.2302 | Yes |
| 205 | ATOX1 | ATOX1 | 21492 | -0.129 | -0.2184 | Yes |
| 206 | FAH | FAH | 21496 | -0.130 | -0.2066 | Yes |
| 207 | PLAU | PLAU | 21507 | -0.141 | -0.1941 | Yes |
| 208 | RHBDF2 | RHBDF2 | 21513 | -0.144 | -0.1810 | Yes |
| 209 | EMP1 | EMP1 | 21515 | -0.145 | -0.1678 | Yes |
| 210 | AHNAK2 | AHNAK2 | 21523 | -0.148 | -0.1545 | Yes |
| 211 | TNFRSF12A | TNFRSF12A | 21534 | -0.154 | -0.1407 | Yes |
| 212 | SERPINE1 | SERPINE1 | 21554 | -0.173 | -0.1257 | Yes |
| 213 | PYGB | PYGB | 21557 | -0.175 | -0.1096 | Yes |
| 214 | PTHLH | PTHLH | 21558 | -0.176 | -0.0934 | Yes |
| 215 | MT1G | MT1G | 21561 | -0.180 | -0.0770 | Yes |
| 216 | IL11 | IL11 | 21567 | -0.185 | -0.0602 | Yes |
| 217 | ITGA3 | ITGA3 | 21577 | -0.205 | -0.0417 | Yes |
| 218 | SLC4A7 | SLC4A7 | 21578 | -0.212 | -0.0222 | Yes |
| 219 | ECM1 | ECM1 | 21591 | -0.249 | 0.0002 | Yes |

