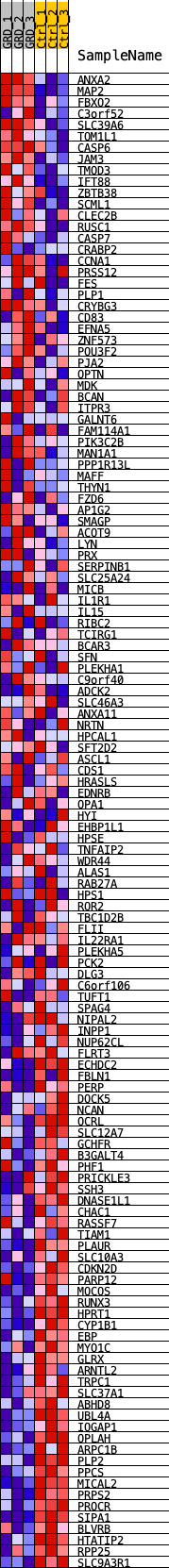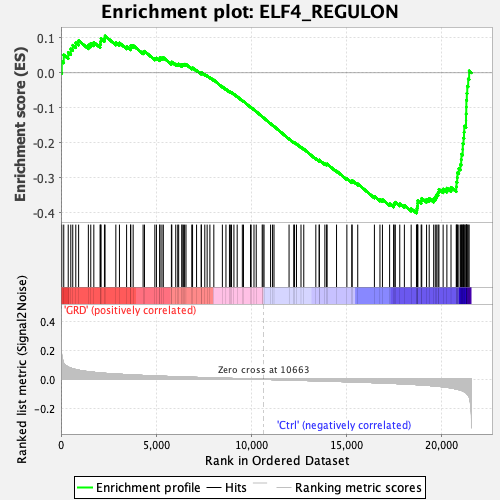
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | ELF4_REGULON |
| Enrichment Score (ES) | -0.40185383 |
| Normalized Enrichment Score (NES) | -1.7274532 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004455574 |
| FWER p-Value | 0.02 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ANXA2 | ANXA2 | 41 | 0.164 | 0.0330 | No |
| 2 | MAP2 | MAP2 | 145 | 0.108 | 0.0514 | No |
| 3 | FBXO2 | FBXO2 | 380 | 0.083 | 0.0583 | No |
| 4 | C3orf52 | C3orf52 | 515 | 0.077 | 0.0685 | No |
| 5 | SLC39A6 | SLC39A6 | 618 | 0.072 | 0.0792 | No |
| 6 | TOM1L1 | TOM1L1 | 778 | 0.067 | 0.0861 | No |
| 7 | CASP6 | CASP6 | 927 | 0.062 | 0.0925 | No |
| 8 | JAM3 | JAM3 | 1439 | 0.052 | 0.0798 | No |
| 9 | TMOD3 | TMOD3 | 1566 | 0.050 | 0.0847 | No |
| 10 | IFT88 | IFT88 | 1727 | 0.048 | 0.0875 | No |
| 11 | ZBTB38 | ZBTB38 | 2056 | 0.044 | 0.0816 | No |
| 12 | SCML1 | SCML1 | 2082 | 0.044 | 0.0898 | No |
| 13 | CLEC2B | CLEC2B | 2106 | 0.043 | 0.0980 | No |
| 14 | RUSC1 | RUSC1 | 2286 | 0.041 | 0.0985 | No |
| 15 | CASP7 | CASP7 | 2317 | 0.041 | 0.1059 | No |
| 16 | CRABP2 | CRABP2 | 2885 | 0.036 | 0.0872 | No |
| 17 | CCNA1 | CCNA1 | 3075 | 0.035 | 0.0859 | No |
| 18 | PRSS12 | PRSS12 | 3451 | 0.032 | 0.0752 | No |
| 19 | FES | FES | 3658 | 0.030 | 0.0721 | No |
| 20 | PLP1 | PLP1 | 3671 | 0.030 | 0.0780 | No |
| 21 | CRYBG3 | CRYBG3 | 3796 | 0.029 | 0.0785 | No |
| 22 | CD83 | CD83 | 4319 | 0.026 | 0.0598 | No |
| 23 | EFNA5 | EFNA5 | 4392 | 0.026 | 0.0620 | No |
| 24 | ZNF573 | ZNF573 | 4937 | 0.023 | 0.0416 | No |
| 25 | POU3F2 | POU3F2 | 5021 | 0.022 | 0.0425 | No |
| 26 | PJA2 | PJA2 | 5193 | 0.022 | 0.0391 | No |
| 27 | OPTN | OPTN | 5205 | 0.022 | 0.0432 | No |
| 28 | MDK | MDK | 5310 | 0.021 | 0.0428 | No |
| 29 | BCAN | BCAN | 5383 | 0.021 | 0.0439 | No |
| 30 | ITPR3 | ITPR3 | 5808 | 0.019 | 0.0281 | No |
| 31 | GALNT6 | GALNT6 | 5829 | 0.019 | 0.0311 | No |
| 32 | FAM114A1 | FAM114A1 | 6037 | 0.018 | 0.0253 | No |
| 33 | PIK3C2B | PIK3C2B | 6142 | 0.017 | 0.0241 | No |
| 34 | MAN1A1 | MAN1A1 | 6191 | 0.017 | 0.0254 | No |
| 35 | PPP1R13L | PPP1R13L | 6359 | 0.016 | 0.0211 | No |
| 36 | MAFF | MAFF | 6367 | 0.016 | 0.0243 | No |
| 37 | THYN1 | THYN1 | 6438 | 0.016 | 0.0244 | No |
| 38 | FZD6 | FZD6 | 6502 | 0.016 | 0.0248 | No |
| 39 | AP1G2 | AP1G2 | 6575 | 0.015 | 0.0247 | No |
| 40 | SMAGP | SMAGP | 6881 | 0.014 | 0.0134 | No |
| 41 | ACOT9 | ACOT9 | 6921 | 0.014 | 0.0146 | No |
| 42 | LYN | LYN | 7132 | 0.013 | 0.0075 | No |
| 43 | PRX | PRX | 7377 | 0.012 | -0.0013 | No |
| 44 | SERPINB1 | SERPINB1 | 7384 | 0.012 | 0.0010 | No |
| 45 | SLC25A24 | SLC25A24 | 7565 | 0.011 | -0.0050 | No |
| 46 | MICB | MICB | 7688 | 0.011 | -0.0083 | No |
| 47 | IL1R1 | IL1R1 | 7833 | 0.010 | -0.0128 | No |
| 48 | IL15 | IL15 | 8040 | 0.009 | -0.0204 | No |
| 49 | RIBC2 | RIBC2 | 8490 | 0.008 | -0.0396 | No |
| 50 | TCIRG1 | TCIRG1 | 8678 | 0.007 | -0.0468 | No |
| 51 | BCAR3 | BCAR3 | 8862 | 0.006 | -0.0540 | No |
| 52 | SFN | SFN | 8909 | 0.006 | -0.0548 | No |
| 53 | PLEKHA1 | PLEKHA1 | 8971 | 0.006 | -0.0563 | No |
| 54 | C9orf40 | C9orf40 | 9097 | 0.006 | -0.0610 | No |
| 55 | ADCK2 | ADCK2 | 9269 | 0.005 | -0.0679 | No |
| 56 | SLC46A3 | SLC46A3 | 9540 | 0.004 | -0.0796 | No |
| 57 | ANXA11 | ANXA11 | 9600 | 0.004 | -0.0815 | No |
| 58 | NRTN | NRTN | 9970 | 0.002 | -0.0982 | No |
| 59 | HPCAL1 | HPCAL1 | 10003 | 0.002 | -0.0992 | No |
| 60 | SFT2D2 | SFT2D2 | 10153 | 0.002 | -0.1057 | No |
| 61 | ASCL1 | ASCL1 | 10271 | 0.001 | -0.1109 | No |
| 62 | CDS1 | CDS1 | 10585 | 0.000 | -0.1254 | No |
| 63 | HRASLS | HRASLS | 10626 | 0.000 | -0.1273 | No |
| 64 | EDNRB | EDNRB | 10682 | -0.000 | -0.1298 | No |
| 65 | OPA1 | OPA1 | 11026 | -0.001 | -0.1455 | No |
| 66 | HYI | HYI | 11130 | -0.002 | -0.1500 | No |
| 67 | EHBP1L1 | EHBP1L1 | 11206 | -0.002 | -0.1531 | No |
| 68 | HPSE | HPSE | 11998 | -0.005 | -0.1890 | No |
| 69 | TNFAIP2 | TNFAIP2 | 12244 | -0.006 | -0.1992 | No |
| 70 | WDR44 | WDR44 | 12287 | -0.006 | -0.1999 | No |
| 71 | ALAS1 | ALAS1 | 12389 | -0.006 | -0.2034 | No |
| 72 | RAB27A | RAB27A | 12626 | -0.007 | -0.2129 | No |
| 73 | HPS1 | HPS1 | 12772 | -0.007 | -0.2181 | No |
| 74 | ROR2 | ROR2 | 13406 | -0.010 | -0.2455 | No |
| 75 | TBC1D2B | TBC1D2B | 13583 | -0.010 | -0.2515 | No |
| 76 | FLII | FLII | 13589 | -0.010 | -0.2495 | No |
| 77 | IL22RA1 | IL22RA1 | 13882 | -0.012 | -0.2606 | No |
| 78 | PLEKHA5 | PLEKHA5 | 13974 | -0.012 | -0.2623 | No |
| 79 | PCK2 | PCK2 | 13998 | -0.012 | -0.2608 | No |
| 80 | DLG3 | DLG3 | 14496 | -0.014 | -0.2810 | No |
| 81 | C6orf106 | C6orf106 | 15039 | -0.016 | -0.3027 | No |
| 82 | TUFT1 | TUFT1 | 15300 | -0.017 | -0.3112 | No |
| 83 | SPAG4 | SPAG4 | 15315 | -0.017 | -0.3081 | No |
| 84 | NIPAL2 | NIPAL2 | 15611 | -0.019 | -0.3179 | No |
| 85 | INPP1 | INPP1 | 16488 | -0.023 | -0.3538 | No |
| 86 | NUP62CL | NUP62CL | 16780 | -0.024 | -0.3622 | No |
| 87 | FLRT3 | FLRT3 | 16911 | -0.025 | -0.3630 | No |
| 88 | ECHDC2 | ECHDC2 | 17288 | -0.027 | -0.3748 | No |
| 89 | FBLN1 | FBLN1 | 17498 | -0.028 | -0.3785 | No |
| 90 | PERP | PERP | 17526 | -0.028 | -0.3737 | No |
| 91 | DOCK5 | DOCK5 | 17588 | -0.029 | -0.3705 | No |
| 92 | NCAN | NCAN | 17822 | -0.030 | -0.3750 | No |
| 93 | OCRL | OCRL | 18065 | -0.031 | -0.3796 | No |
| 94 | SLC12A7 | SLC12A7 | 18422 | -0.034 | -0.3889 | No |
| 95 | GCHFR | GCHFR | 18701 | -0.036 | -0.3941 | Yes |
| 96 | B3GALT4 | B3GALT4 | 18726 | -0.036 | -0.3875 | Yes |
| 97 | PHF1 | PHF1 | 18754 | -0.036 | -0.3810 | Yes |
| 98 | PRICKLE3 | PRICKLE3 | 18763 | -0.037 | -0.3735 | Yes |
| 99 | SSH3 | SSH3 | 18771 | -0.037 | -0.3660 | Yes |
| 100 | DNASE1L1 | DNASE1L1 | 18949 | -0.038 | -0.3661 | Yes |
| 101 | CHAC1 | CHAC1 | 18979 | -0.038 | -0.3592 | Yes |
| 102 | RASSF7 | RASSF7 | 19232 | -0.041 | -0.3622 | Yes |
| 103 | TIAM1 | TIAM1 | 19371 | -0.042 | -0.3597 | Yes |
| 104 | PLAUR | PLAUR | 19606 | -0.045 | -0.3610 | Yes |
| 105 | SLC10A3 | SLC10A3 | 19691 | -0.046 | -0.3552 | Yes |
| 106 | CDKN2D | CDKN2D | 19768 | -0.047 | -0.3487 | Yes |
| 107 | PARP12 | PARP12 | 19840 | -0.048 | -0.3418 | Yes |
| 108 | MOCOS | MOCOS | 19883 | -0.048 | -0.3335 | Yes |
| 109 | RUNX3 | RUNX3 | 20104 | -0.051 | -0.3327 | Yes |
| 110 | HPRT1 | HPRT1 | 20301 | -0.055 | -0.3302 | Yes |
| 111 | CYP1B1 | CYP1B1 | 20518 | -0.059 | -0.3277 | Yes |
| 112 | EBP | EBP | 20794 | -0.066 | -0.3264 | Yes |
| 113 | MYO1C | MYO1C | 20807 | -0.066 | -0.3128 | Yes |
| 114 | GLRX | GLRX | 20844 | -0.068 | -0.3000 | Yes |
| 115 | ARNTL2 | ARNTL2 | 20855 | -0.068 | -0.2859 | Yes |
| 116 | TRPC1 | TRPC1 | 20930 | -0.070 | -0.2744 | Yes |
| 117 | SLC37A1 | SLC37A1 | 21013 | -0.073 | -0.2625 | Yes |
| 118 | ABHD8 | ABHD8 | 21053 | -0.075 | -0.2484 | Yes |
| 119 | UBL4A | UBL4A | 21067 | -0.075 | -0.2328 | Yes |
| 120 | IQGAP1 | IQGAP1 | 21134 | -0.079 | -0.2191 | Yes |
| 121 | OPLAH | OPLAH | 21138 | -0.079 | -0.2023 | Yes |
| 122 | ARPC1B | ARPC1B | 21177 | -0.082 | -0.1866 | Yes |
| 123 | PLP2 | PLP2 | 21203 | -0.083 | -0.1699 | Yes |
| 124 | PPCS | PPCS | 21215 | -0.084 | -0.1524 | Yes |
| 125 | MICAL2 | MICAL2 | 21308 | -0.091 | -0.1371 | Yes |
| 126 | PRPS2 | PRPS2 | 21310 | -0.092 | -0.1176 | Yes |
| 127 | PROCR | PROCR | 21321 | -0.093 | -0.0981 | Yes |
| 128 | SIPA1 | SIPA1 | 21326 | -0.094 | -0.0782 | Yes |
| 129 | BLVRB | BLVRB | 21350 | -0.098 | -0.0584 | Yes |
| 130 | HTATIP2 | HTATIP2 | 21373 | -0.101 | -0.0379 | Yes |
| 131 | RPP25 | RPP25 | 21422 | -0.107 | -0.0173 | Yes |
| 132 | SLC9A3R1 | SLC9A3R1 | 21472 | -0.118 | 0.0057 | Yes |