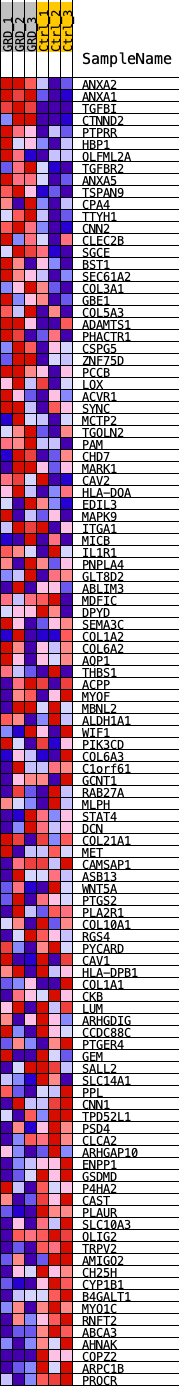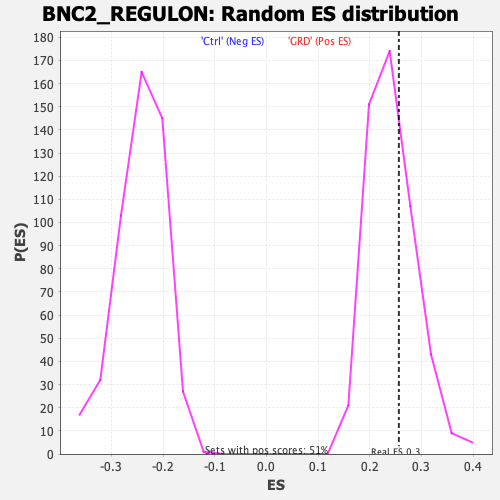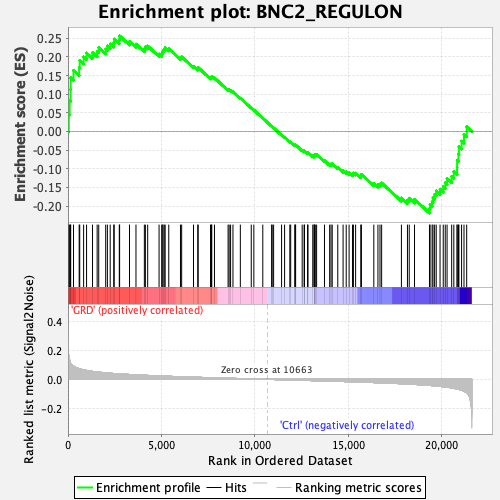
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | GRD |
| GeneSet | BNC2_REGULON |
| Enrichment Score (ES) | 0.25640944 |
| Normalized Enrichment Score (NES) | 1.0574288 |
| Nominal p-value | 0.33529413 |
| FDR q-value | 0.32299724 |
| FWER p-Value | 0.997 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ANXA2 | ANXA2 | 41 | 0.164 | 0.0458 | Yes |
| 2 | ANXA1 | ANXA1 | 87 | 0.133 | 0.0827 | Yes |
| 3 | TGFBI | TGFBI | 137 | 0.111 | 0.1127 | Yes |
| 4 | CTNND2 | CTNND2 | 142 | 0.109 | 0.1444 | Yes |
| 5 | PTPRR | PTPRR | 295 | 0.091 | 0.1639 | Yes |
| 6 | HBP1 | HBP1 | 597 | 0.073 | 0.1712 | Yes |
| 7 | OLFML2A | OLFML2A | 627 | 0.072 | 0.1908 | Yes |
| 8 | TGFBR2 | TGFBR2 | 835 | 0.065 | 0.2000 | Yes |
| 9 | ANXA5 | ANXA5 | 993 | 0.061 | 0.2104 | Yes |
| 10 | TSPAN9 | TSPAN9 | 1311 | 0.054 | 0.2116 | Yes |
| 11 | CPA4 | CPA4 | 1559 | 0.050 | 0.2148 | Yes |
| 12 | TTYH1 | TTYH1 | 1643 | 0.049 | 0.2253 | Yes |
| 13 | CNN2 | CNN2 | 2016 | 0.045 | 0.2210 | Yes |
| 14 | CLEC2B | CLEC2B | 2106 | 0.043 | 0.2295 | Yes |
| 15 | SGCE | SGCE | 2256 | 0.042 | 0.2348 | Yes |
| 16 | BST1 | BST1 | 2447 | 0.040 | 0.2375 | Yes |
| 17 | SEC61A2 | SEC61A2 | 2469 | 0.040 | 0.2481 | Yes |
| 18 | COL3A1 | COL3A1 | 2743 | 0.037 | 0.2463 | Yes |
| 19 | GBE1 | GBE1 | 2760 | 0.037 | 0.2564 | Yes |
| 20 | COL5A3 | COL5A3 | 3289 | 0.033 | 0.2415 | No |
| 21 | ADAMTS1 | ADAMTS1 | 3635 | 0.031 | 0.2344 | No |
| 22 | PHACTR1 | PHACTR1 | 4083 | 0.028 | 0.2217 | No |
| 23 | CSPG5 | CSPG5 | 4138 | 0.027 | 0.2272 | No |
| 24 | ZNF75D | ZNF75D | 4261 | 0.027 | 0.2293 | No |
| 25 | PCCB | PCCB | 4870 | 0.023 | 0.2077 | No |
| 26 | LOX | LOX | 5014 | 0.022 | 0.2076 | No |
| 27 | ACVR1 | ACVR1 | 5036 | 0.022 | 0.2132 | No |
| 28 | SYNC | SYNC | 5085 | 0.022 | 0.2174 | No |
| 29 | MCTP2 | MCTP2 | 5147 | 0.022 | 0.2209 | No |
| 30 | TGOLN2 | TGOLN2 | 5196 | 0.022 | 0.2249 | No |
| 31 | PAM | PAM | 5389 | 0.021 | 0.2220 | No |
| 32 | CHD7 | CHD7 | 6015 | 0.018 | 0.1981 | No |
| 33 | MARK1 | MARK1 | 6078 | 0.017 | 0.2003 | No |
| 34 | CAV2 | CAV2 | 6711 | 0.015 | 0.1751 | No |
| 35 | HLA-DOA | HLA-DOA | 6941 | 0.014 | 0.1685 | No |
| 36 | EDIL3 | EDIL3 | 6960 | 0.014 | 0.1716 | No |
| 37 | MAPK9 | MAPK9 | 7622 | 0.011 | 0.1441 | No |
| 38 | ITGA1 | ITGA1 | 7671 | 0.011 | 0.1450 | No |
| 39 | MICB | MICB | 7688 | 0.011 | 0.1474 | No |
| 40 | IL1R1 | IL1R1 | 7833 | 0.010 | 0.1438 | No |
| 41 | PNPLA4 | PNPLA4 | 8561 | 0.008 | 0.1122 | No |
| 42 | GLT8D2 | GLT8D2 | 8617 | 0.007 | 0.1118 | No |
| 43 | ABLIM3 | ABLIM3 | 8694 | 0.007 | 0.1103 | No |
| 44 | MDFIC | MDFIC | 8822 | 0.007 | 0.1063 | No |
| 45 | DPYD | DPYD | 9216 | 0.005 | 0.0895 | No |
| 46 | SEMA3C | SEMA3C | 9800 | 0.003 | 0.0633 | No |
| 47 | COL1A2 | COL1A2 | 9939 | 0.003 | 0.0576 | No |
| 48 | COL6A2 | COL6A2 | 10417 | 0.001 | 0.0356 | No |
| 49 | AQP1 | AQP1 | 10889 | -0.001 | 0.0139 | No |
| 50 | THBS1 | THBS1 | 10919 | -0.001 | 0.0128 | No |
| 51 | ACPP | ACPP | 10996 | -0.001 | 0.0096 | No |
| 52 | MYOF | MYOF | 11422 | -0.003 | -0.0094 | No |
| 53 | MBNL2 | MBNL2 | 11583 | -0.003 | -0.0159 | No |
| 54 | ALDH1A1 | ALDH1A1 | 11863 | -0.004 | -0.0277 | No |
| 55 | WIF1 | WIF1 | 11909 | -0.004 | -0.0285 | No |
| 56 | PIK3CD | PIK3CD | 12129 | -0.005 | -0.0372 | No |
| 57 | COL6A3 | COL6A3 | 12151 | -0.005 | -0.0367 | No |
| 58 | C1orf61 | C1orf61 | 12173 | -0.005 | -0.0361 | No |
| 59 | GCNT1 | GCNT1 | 12523 | -0.007 | -0.0505 | No |
| 60 | RAB27A | RAB27A | 12626 | -0.007 | -0.0532 | No |
| 61 | MLPH | MLPH | 12647 | -0.007 | -0.0521 | No |
| 62 | STAT4 | STAT4 | 12813 | -0.008 | -0.0576 | No |
| 63 | DCN | DCN | 12833 | -0.008 | -0.0563 | No |
| 64 | COL21A1 | COL21A1 | 13090 | -0.009 | -0.0657 | No |
| 65 | MET | MET | 13160 | -0.009 | -0.0663 | No |
| 66 | CAMSAP1 | CAMSAP1 | 13168 | -0.009 | -0.0640 | No |
| 67 | ASB13 | ASB13 | 13194 | -0.009 | -0.0626 | No |
| 68 | WNT5A | WNT5A | 13222 | -0.009 | -0.0612 | No |
| 69 | PTGS2 | PTGS2 | 13295 | -0.009 | -0.0618 | No |
| 70 | PLA2R1 | PLA2R1 | 13716 | -0.011 | -0.0782 | No |
| 71 | COL10A1 | COL10A1 | 13993 | -0.012 | -0.0875 | No |
| 72 | RGS4 | RGS4 | 14065 | -0.012 | -0.0872 | No |
| 73 | PYCARD | PYCARD | 14130 | -0.013 | -0.0865 | No |
| 74 | CAV1 | CAV1 | 14427 | -0.014 | -0.0963 | No |
| 75 | HLA-DPB1 | HLA-DPB1 | 14711 | -0.015 | -0.1051 | No |
| 76 | COL1A1 | COL1A1 | 14878 | -0.016 | -0.1083 | No |
| 77 | CKB | CKB | 15032 | -0.016 | -0.1107 | No |
| 78 | LUM | LUM | 15213 | -0.017 | -0.1142 | No |
| 79 | ARHGDIG | ARHGDIG | 15263 | -0.017 | -0.1114 | No |
| 80 | CCDC88C | CCDC88C | 15381 | -0.018 | -0.1117 | No |
| 81 | PTGER4 | PTGER4 | 15657 | -0.019 | -0.1190 | No |
| 82 | GEM | GEM | 15692 | -0.019 | -0.1150 | No |
| 83 | SALL2 | SALL2 | 16354 | -0.022 | -0.1394 | No |
| 84 | SLC14A1 | SLC14A1 | 16575 | -0.023 | -0.1429 | No |
| 85 | PPL | PPL | 16685 | -0.024 | -0.1410 | No |
| 86 | CNN1 | CNN1 | 16773 | -0.024 | -0.1380 | No |
| 87 | TPD52L1 | TPD52L1 | 17830 | -0.030 | -0.1785 | No |
| 88 | PSD4 | PSD4 | 18153 | -0.032 | -0.1841 | No |
| 89 | CLCA2 | CLCA2 | 18248 | -0.033 | -0.1790 | No |
| 90 | ARHGAP10 | ARHGAP10 | 18531 | -0.035 | -0.1819 | No |
| 91 | ENPP1 | ENPP1 | 19331 | -0.042 | -0.2069 | No |
| 92 | GSDMD | GSDMD | 19363 | -0.042 | -0.1961 | No |
| 93 | P4HA2 | P4HA2 | 19472 | -0.043 | -0.1885 | No |
| 94 | CAST | CAST | 19519 | -0.044 | -0.1779 | No |
| 95 | PLAUR | PLAUR | 19606 | -0.045 | -0.1688 | No |
| 96 | SLC10A3 | SLC10A3 | 19691 | -0.046 | -0.1594 | No |
| 97 | OLIG2 | OLIG2 | 19896 | -0.049 | -0.1547 | No |
| 98 | TRPV2 | TRPV2 | 20067 | -0.051 | -0.1478 | No |
| 99 | AMIGO2 | AMIGO2 | 20174 | -0.052 | -0.1374 | No |
| 100 | CH25H | CH25H | 20272 | -0.054 | -0.1261 | No |
| 101 | CYP1B1 | CYP1B1 | 20518 | -0.059 | -0.1205 | No |
| 102 | B4GALT1 | B4GALT1 | 20633 | -0.062 | -0.1078 | No |
| 103 | MYO1C | MYO1C | 20807 | -0.066 | -0.0964 | No |
| 104 | RNFT2 | RNFT2 | 20811 | -0.067 | -0.0771 | No |
| 105 | ABCA3 | ABCA3 | 20881 | -0.069 | -0.0603 | No |
| 106 | AHNAK | AHNAK | 20906 | -0.070 | -0.0410 | No |
| 107 | COPZ2 | COPZ2 | 21048 | -0.075 | -0.0258 | No |
| 108 | ARPC1B | ARPC1B | 21177 | -0.082 | -0.0079 | No |
| 109 | PROCR | PROCR | 21321 | -0.093 | 0.0128 | No |