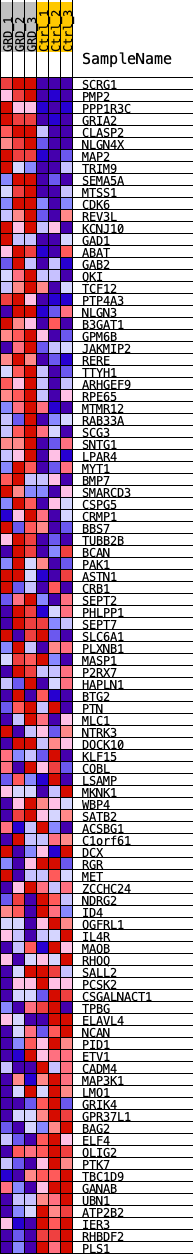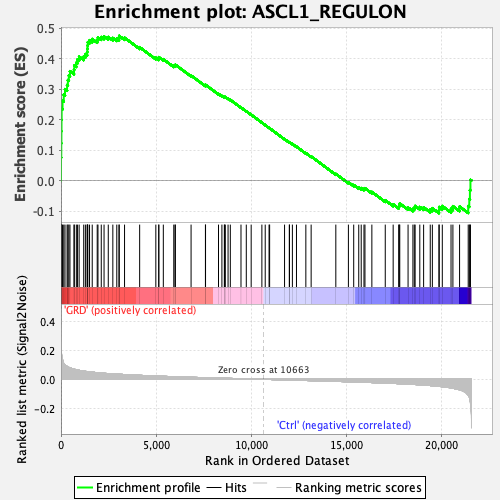
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | GRD |
| GeneSet | ASCL1_REGULON |
| Enrichment Score (ES) | 0.47528905 |
| Normalized Enrichment Score (NES) | 1.9207528 |
| Nominal p-value | 0.0 |
| FDR q-value | 7.443609E-4 |
| FWER p-Value | 0.002 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SCRG1 | SCRG1 | 1 | 0.360 | 0.0762 | Yes |
| 2 | PMP2 | PMP2 | 15 | 0.222 | 0.1225 | Yes |
| 3 | PPP1R3C | PPP1R3C | 22 | 0.188 | 0.1621 | Yes |
| 4 | GRIA2 | GRIA2 | 27 | 0.178 | 0.1997 | Yes |
| 5 | CLASP2 | CLASP2 | 34 | 0.171 | 0.2357 | Yes |
| 6 | NLGN4X | NLGN4X | 81 | 0.135 | 0.2622 | Yes |
| 7 | MAP2 | MAP2 | 145 | 0.108 | 0.2822 | Yes |
| 8 | TRIM9 | TRIM9 | 210 | 0.099 | 0.3002 | Yes |
| 9 | SEMA5A | SEMA5A | 320 | 0.089 | 0.3139 | Yes |
| 10 | MTSS1 | MTSS1 | 362 | 0.085 | 0.3300 | Yes |
| 11 | CDK6 | CDK6 | 413 | 0.082 | 0.3450 | Yes |
| 12 | REV3L | REV3L | 471 | 0.079 | 0.3591 | Yes |
| 13 | KCNJ10 | KCNJ10 | 684 | 0.070 | 0.3641 | Yes |
| 14 | GAD1 | GAD1 | 699 | 0.069 | 0.3782 | Yes |
| 15 | ABAT | ABAT | 806 | 0.066 | 0.3871 | Yes |
| 16 | GAB2 | GAB2 | 865 | 0.064 | 0.3980 | Yes |
| 17 | QKI | QKI | 954 | 0.062 | 0.4070 | Yes |
| 18 | TCF12 | TCF12 | 1195 | 0.057 | 0.4078 | Yes |
| 19 | PTP4A3 | PTP4A3 | 1283 | 0.055 | 0.4153 | Yes |
| 20 | NLGN3 | NLGN3 | 1376 | 0.053 | 0.4223 | Yes |
| 21 | B3GAT1 | B3GAT1 | 1387 | 0.053 | 0.4330 | Yes |
| 22 | GPM6B | GPM6B | 1398 | 0.053 | 0.4437 | Yes |
| 23 | JAKMIP2 | JAKMIP2 | 1409 | 0.053 | 0.4544 | Yes |
| 24 | RERE | RERE | 1501 | 0.051 | 0.4610 | Yes |
| 25 | TTYH1 | TTYH1 | 1643 | 0.049 | 0.4648 | Yes |
| 26 | ARHGEF9 | ARHGEF9 | 1905 | 0.046 | 0.4624 | Yes |
| 27 | RPE65 | RPE65 | 1945 | 0.045 | 0.4702 | Yes |
| 28 | MTMR12 | MTMR12 | 2118 | 0.043 | 0.4714 | Yes |
| 29 | RAB33A | RAB33A | 2265 | 0.042 | 0.4734 | Yes |
| 30 | SCG3 | SCG3 | 2487 | 0.039 | 0.4715 | Yes |
| 31 | SNTG1 | SNTG1 | 2727 | 0.038 | 0.4683 | Yes |
| 32 | LPAR4 | LPAR4 | 2935 | 0.036 | 0.4663 | Yes |
| 33 | MYT1 | MYT1 | 3053 | 0.035 | 0.4682 | Yes |
| 34 | BMP7 | BMP7 | 3061 | 0.035 | 0.4753 | Yes |
| 35 | SMARCD3 | SMARCD3 | 3343 | 0.033 | 0.4691 | No |
| 36 | CSPG5 | CSPG5 | 4138 | 0.027 | 0.4380 | No |
| 37 | CRMP1 | CRMP1 | 4989 | 0.023 | 0.4032 | No |
| 38 | BBS7 | BBS7 | 5144 | 0.022 | 0.4007 | No |
| 39 | TUBB2B | TUBB2B | 5145 | 0.022 | 0.4053 | No |
| 40 | BCAN | BCAN | 5383 | 0.021 | 0.3986 | No |
| 41 | PAK1 | PAK1 | 5933 | 0.018 | 0.3769 | No |
| 42 | ASTN1 | ASTN1 | 5998 | 0.018 | 0.3777 | No |
| 43 | CRB1 | CRB1 | 6017 | 0.018 | 0.3806 | No |
| 44 | SEPT2 | SEPT2 | 6843 | 0.014 | 0.3452 | No |
| 45 | PHLPP1 | PHLPP1 | 7598 | 0.011 | 0.3125 | No |
| 46 | SEPT7 | SEPT7 | 7604 | 0.011 | 0.3146 | No |
| 47 | SLC6A1 | SLC6A1 | 8286 | 0.009 | 0.2847 | No |
| 48 | PLXNB1 | PLXNB1 | 8465 | 0.008 | 0.2781 | No |
| 49 | MASP1 | MASP1 | 8593 | 0.007 | 0.2738 | No |
| 50 | P2RX7 | P2RX7 | 8606 | 0.007 | 0.2748 | No |
| 51 | HAPLN1 | HAPLN1 | 8644 | 0.007 | 0.2746 | No |
| 52 | BTG2 | BTG2 | 8789 | 0.007 | 0.2694 | No |
| 53 | PTN | PTN | 8908 | 0.006 | 0.2652 | No |
| 54 | MLC1 | MLC1 | 9469 | 0.004 | 0.2401 | No |
| 55 | NTRK3 | NTRK3 | 9748 | 0.003 | 0.2278 | No |
| 56 | DOCK10 | DOCK10 | 10005 | 0.002 | 0.2164 | No |
| 57 | KLF15 | KLF15 | 10568 | 0.000 | 0.1903 | No |
| 58 | COBL | COBL | 10744 | -0.000 | 0.1822 | No |
| 59 | LSAMP | LSAMP | 10946 | -0.001 | 0.1731 | No |
| 60 | MKNK1 | MKNK1 | 10976 | -0.001 | 0.1719 | No |
| 61 | WBP4 | WBP4 | 11761 | -0.004 | 0.1363 | No |
| 62 | SATB2 | SATB2 | 12011 | -0.005 | 0.1257 | No |
| 63 | ACSBG1 | ACSBG1 | 12021 | -0.005 | 0.1263 | No |
| 64 | C1orf61 | C1orf61 | 12173 | -0.005 | 0.1203 | No |
| 65 | DCX | DCX | 12388 | -0.006 | 0.1117 | No |
| 66 | RGR | RGR | 12881 | -0.008 | 0.0904 | No |
| 67 | MET | MET | 13160 | -0.009 | 0.0794 | No |
| 68 | ZCCHC24 | ZCCHC24 | 14458 | -0.014 | 0.0220 | No |
| 69 | NDRG2 | NDRG2 | 15120 | -0.017 | -0.0053 | No |
| 70 | ID4 | ID4 | 15399 | -0.018 | -0.0144 | No |
| 71 | OGFRL1 | OGFRL1 | 15664 | -0.019 | -0.0227 | No |
| 72 | IL4R | IL4R | 15794 | -0.020 | -0.0246 | No |
| 73 | MAOB | MAOB | 15939 | -0.020 | -0.0270 | No |
| 74 | RHOQ | RHOQ | 15991 | -0.020 | -0.0250 | No |
| 75 | SALL2 | SALL2 | 16354 | -0.022 | -0.0372 | No |
| 76 | PCSK2 | PCSK2 | 17059 | -0.026 | -0.0645 | No |
| 77 | CSGALNACT1 | CSGALNACT1 | 17475 | -0.028 | -0.0779 | No |
| 78 | TPBG | TPBG | 17767 | -0.029 | -0.0852 | No |
| 79 | ELAVL4 | ELAVL4 | 17794 | -0.030 | -0.0802 | No |
| 80 | NCAN | NCAN | 17822 | -0.030 | -0.0751 | No |
| 81 | PID1 | PID1 | 18259 | -0.033 | -0.0885 | No |
| 82 | ETV1 | ETV1 | 18514 | -0.035 | -0.0930 | No |
| 83 | CADM4 | CADM4 | 18586 | -0.035 | -0.0888 | No |
| 84 | MAP3K1 | MAP3K1 | 18630 | -0.035 | -0.0833 | No |
| 85 | LMO1 | LMO1 | 18876 | -0.038 | -0.0867 | No |
| 86 | GRIK4 | GRIK4 | 19077 | -0.039 | -0.0877 | No |
| 87 | GPR37L1 | GPR37L1 | 19426 | -0.043 | -0.0948 | No |
| 88 | BAG2 | BAG2 | 19537 | -0.044 | -0.0906 | No |
| 89 | ELF4 | ELF4 | 19885 | -0.048 | -0.0966 | No |
| 90 | OLIG2 | OLIG2 | 19896 | -0.049 | -0.0867 | No |
| 91 | PTK7 | PTK7 | 20060 | -0.051 | -0.0836 | No |
| 92 | TBC1D9 | TBC1D9 | 20519 | -0.059 | -0.0925 | No |
| 93 | GANAB | GANAB | 20618 | -0.061 | -0.0841 | No |
| 94 | UBN1 | UBN1 | 20969 | -0.072 | -0.0852 | No |
| 95 | ATP2B2 | ATP2B2 | 21423 | -0.107 | -0.0836 | No |
| 96 | IER3 | IER3 | 21480 | -0.122 | -0.0604 | No |
| 97 | RHBDF2 | RHBDF2 | 21513 | -0.144 | -0.0313 | No |
| 98 | PLS1 | PLS1 | 21541 | -0.166 | 0.0025 | No |