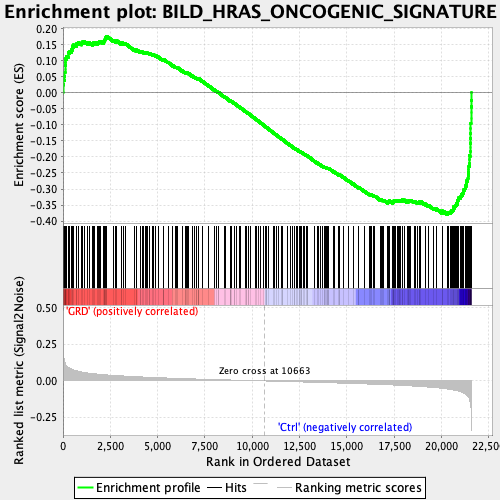
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | NF1_GRD_exprs.NF1_GRD_phenotype.cls #GRD_versus_Ctrl.NF1_GRD_phenotype.cls #GRD_versus_Ctrl_repos |
| Phenotype | NF1_GRD_phenotype.cls#GRD_versus_Ctrl_repos |
| Upregulated in class | Ctrl |
| GeneSet | BILD_HRAS_ONCOGENIC_SIGNATURE |
| Enrichment Score (ES) | -0.37952545 |
| Normalized Enrichment Score (NES) | -1.761186 |
| Nominal p-value | 0.0 |
| FDR q-value | 5.3571427E-4 |
| FWER p-Value | 0.002 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | VEGFA | VEGFA | 2 | 0.280 | 0.0259 | No | ||
| 2 | SLC16A3 | SLC16A3 | 31 | 0.173 | 0.0407 | No | ||
| 3 | FLNB | FLNB | 64 | 0.143 | 0.0524 | No | ||
| 4 | KLF6 | KLF6 | 97 | 0.128 | 0.0629 | No | ||
| 5 | STAG3L1 | STAG3L1 | 103 | 0.125 | 0.0742 | No | ||
| 6 | FOS | FOS | 104 | 0.125 | 0.0858 | No | ||
| 7 | TPM1 | TPM1 | 120 | 0.116 | 0.0958 | No | ||
| 8 | ZNF608 | ZNF608 | 125 | 0.113 | 0.1061 | No | ||
| 9 | HNRNPH1 | HNRNPH1 | 173 | 0.104 | 0.1136 | No | ||
| 10 | MEG3 | MEG3 | 268 | 0.093 | 0.1178 | No | ||
| 11 | CCNL1 | CCNL1 | 269 | 0.093 | 0.1264 | No | ||
| 12 | EPHA4 | EPHA4 | 356 | 0.086 | 0.1303 | No | ||
| 13 | NAMPT | NAMPT | 433 | 0.081 | 0.1343 | No | ||
| 14 | PPP1R15A | PPP1R15A | 477 | 0.079 | 0.1396 | No | ||
| 15 | KLF5 | KLF5 | 486 | 0.078 | 0.1465 | No | ||
| 16 | ZNF827 | ZNF827 | 564 | 0.074 | 0.1498 | No | ||
| 17 | UPP1 | UPP1 | 694 | 0.070 | 0.1502 | No | ||
| 18 | DUSP4 | DUSP4 | 727 | 0.068 | 0.1550 | No | ||
| 19 | GLTP | GLTP | 801 | 0.066 | 0.1577 | No | ||
| 20 | SLC2A3 | SLC2A3 | 977 | 0.061 | 0.1552 | No | ||
| 21 | SOCS1 | SOCS1 | 1015 | 0.060 | 0.1590 | No | ||
| 22 | PALMD | PALMD | 1113 | 0.058 | 0.1599 | No | ||
| 23 | ANGPTL4 | ANGPTL4 | 1289 | 0.055 | 0.1568 | No | ||
| 24 | PVR | PVR | 1393 | 0.053 | 0.1569 | No | ||
| 25 | AKAP12 | AKAP12 | 1567 | 0.050 | 0.1534 | No | ||
| 26 | CASP2 | CASP2 | 1608 | 0.050 | 0.1562 | No | ||
| 27 | JUNB | JUNB | 1660 | 0.049 | 0.1583 | No | ||
| 28 | TIA1 | TIA1 | 1816 | 0.047 | 0.1554 | No | ||
| 29 | VANGL2 | VANGL2 | 1851 | 0.046 | 0.1581 | No | ||
| 30 | ARHGEF9 | ARHGEF9 | 1905 | 0.046 | 0.1599 | No | ||
| 31 | BCL6 | BCL6 | 1952 | 0.045 | 0.1619 | No | ||
| 32 | SEMA4B | SEMA4B | 2149 | 0.043 | 0.1568 | No | ||
| 33 | MID1 | MID1 | 2162 | 0.043 | 0.1602 | No | ||
| 34 | HIST1H2AC | HIST1H2AC | 2186 | 0.043 | 0.1630 | No | ||
| 35 | C6orf141 | C6orf141 | 2212 | 0.042 | 0.1658 | No | ||
| 36 | LRIG3 | LRIG3 | 2227 | 0.042 | 0.1690 | No | ||
| 37 | MALAT1 | MALAT1 | 2253 | 0.042 | 0.1717 | No | ||
| 38 | NOLC1 | NOLC1 | 2279 | 0.041 | 0.1744 | No | ||
| 39 | TBX3 | TBX3 | 2301 | 0.041 | 0.1773 | No | ||
| 40 | DDX17 | DDX17 | 2679 | 0.038 | 0.1631 | No | ||
| 41 | HOXC6 | HOXC6 | 2789 | 0.037 | 0.1614 | No | ||
| 42 | XIST | XIST | 2824 | 0.037 | 0.1632 | No | ||
| 43 | CCNA1 | CCNA1 | 3075 | 0.035 | 0.1548 | No | ||
| 44 | PTX3 | PTX3 | 3107 | 0.034 | 0.1565 | No | ||
| 45 | MLL3 | MLL3 | 3206 | 0.034 | 0.1550 | No | ||
| 46 | ITGA2 | ITGA2 | 3297 | 0.033 | 0.1539 | No | ||
| 47 | RRP9 | RRP9 | 3751 | 0.030 | 0.1354 | No | ||
| 48 | OXSR1 | OXSR1 | 3867 | 0.029 | 0.1327 | No | ||
| 49 | MYD88 | MYD88 | 3892 | 0.029 | 0.1343 | No | ||
| 50 | ST5 | ST5 | 4088 | 0.028 | 0.1277 | No | ||
| 51 | ELOVL7 | ELOVL7 | 4111 | 0.028 | 0.1292 | No | ||
| 52 | NIPAL1 | NIPAL1 | 4192 | 0.027 | 0.1280 | No | ||
| 53 | ADRB2 | ADRB2 | 4272 | 0.027 | 0.1268 | No | ||
| 54 | KIAA0528 | KIAA0528 | 4366 | 0.026 | 0.1248 | No | ||
| 55 | HIST1H1E | HIST1H1E | 4382 | 0.026 | 0.1265 | No | ||
| 56 | GLCCI1 | GLCCI1 | 4470 | 0.025 | 0.1248 | No | ||
| 57 | TNFRSF10A | TNFRSF10A | 4553 | 0.025 | 0.1233 | No | ||
| 58 | NFIB | NFIB | 4586 | 0.025 | 0.1241 | No | ||
| 59 | ZFP36 | ZFP36 | 4748 | 0.024 | 0.1188 | No | ||
| 60 | HRAS | HRAS | 4781 | 0.024 | 0.1195 | No | ||
| 61 | TGFA | TGFA | 4900 | 0.023 | 0.1161 | No | ||
| 62 | PNLIPRP3 | PNLIPRP3 | 5025 | 0.022 | 0.1123 | No | ||
| 63 | DNAJC6 | DNAJC6 | 5289 | 0.021 | 0.1020 | No | ||
| 64 | ELK3 | ELK3 | 5298 | 0.021 | 0.1035 | No | ||
| 65 | MTPAP | MTPAP | 5314 | 0.021 | 0.1048 | No | ||
| 66 | PNMA2 | PNMA2 | 5558 | 0.020 | 0.0952 | No | ||
| 67 | ITPR3 | ITPR3 | 5808 | 0.019 | 0.0853 | No | ||
| 68 | RNF152 | RNF152 | 5951 | 0.018 | 0.0803 | No | ||
| 69 | TNRC6A | TNRC6A | 5993 | 0.018 | 0.0800 | No | ||
| 70 | ZBED2 | ZBED2 | 6053 | 0.017 | 0.0789 | No | ||
| 71 | SERPINB2 | SERPINB2 | 6290 | 0.016 | 0.0694 | No | ||
| 72 | CSNK1E | CSNK1E | 6457 | 0.016 | 0.0630 | No | ||
| 73 | EFNB1 | EFNB1 | 6468 | 0.016 | 0.0640 | No | ||
| 74 | MBOAT2 | MBOAT2 | 6497 | 0.016 | 0.0642 | No | ||
| 75 | MED25 | MED25 | 6554 | 0.015 | 0.0630 | No | ||
| 76 | MAST4 | MAST4 | 6641 | 0.015 | 0.0603 | No | ||
| 77 | TMTC3 | TMTC3 | 6826 | 0.014 | 0.0530 | No | ||
| 78 | LRAT | LRAT | 6935 | 0.014 | 0.0492 | No | ||
| 79 | KCNK1 | KCNK1 | 7034 | 0.013 | 0.0459 | No | ||
| 80 | NAV3 | NAV3 | 7054 | 0.013 | 0.0462 | No | ||
| 81 | LYN | LYN | 7132 | 0.013 | 0.0438 | No | ||
| 82 | FN1 | FN1 | 7138 | 0.013 | 0.0448 | No | ||
| 83 | HK2 | HK2 | 7149 | 0.013 | 0.0455 | No | ||
| 84 | SERPINB1 | SERPINB1 | 7384 | 0.012 | 0.0356 | No | ||
| 85 | TOR1AIP1 | TOR1AIP1 | 7699 | 0.011 | 0.0219 | No | ||
| 86 | FOXQ1 | FOXQ1 | 7982 | 0.010 | 0.0096 | No | ||
| 87 | IL8 | IL8 | 8099 | 0.009 | 0.0050 | No | ||
| 88 | KDM6B | KDM6B | 8195 | 0.009 | 0.0014 | No | ||
| 89 | CALM2 | CALM2 | 8218 | 0.009 | 0.0012 | No | ||
| 90 | TOB1 | TOB1 | 8531 | 0.008 | -0.0127 | No | ||
| 91 | TTC8 | TTC8 | 8582 | 0.008 | -0.0144 | No | ||
| 92 | TOP1 | TOP1 | 8831 | 0.007 | -0.0254 | No | ||
| 93 | PIM1 | PIM1 | 8834 | 0.006 | -0.0249 | No | ||
| 94 | DUSP1 | DUSP1 | 8850 | 0.006 | -0.0250 | No | ||
| 95 | SFN | SFN | 8909 | 0.006 | -0.0271 | No | ||
| 96 | ACOT7 | ACOT7 | 9041 | 0.006 | -0.0327 | No | ||
| 97 | PDGFA | PDGFA | 9046 | 0.006 | -0.0323 | No | ||
| 98 | GJB3 | GJB3 | 9178 | 0.005 | -0.0380 | No | ||
| 99 | CSNK1D | CSNK1D | 9324 | 0.005 | -0.0443 | No | ||
| 100 | CDCP1 | CDCP1 | 9398 | 0.005 | -0.0473 | No | ||
| 101 | IRX2 | IRX2 | 9631 | 0.004 | -0.0579 | No | ||
| 102 | ADAMTS5 | ADAMTS5 | 9676 | 0.003 | -0.0596 | No | ||
| 103 | SERPINB3 | SERPINB3 | 9711 | 0.003 | -0.0609 | No | ||
| 104 | ZNF192 | ZNF192 | 9786 | 0.003 | -0.0641 | No | ||
| 105 | SPRY4 | SPRY4 | 9820 | 0.003 | -0.0654 | No | ||
| 106 | PIAS1 | PIAS1 | 9931 | 0.003 | -0.0703 | No | ||
| 107 | IER2 | IER2 | 10164 | 0.002 | -0.0810 | No | ||
| 108 | SAMD8 | SAMD8 | 10199 | 0.002 | -0.0824 | No | ||
| 109 | KCNN4 | KCNN4 | 10334 | 0.001 | -0.0886 | No | ||
| 110 | LGALS8 | LGALS8 | 10438 | 0.001 | -0.0933 | No | ||
| 111 | COL27A1 | COL27A1 | 10591 | 0.000 | -0.1004 | No | ||
| 112 | ABCA1 | ABCA1 | 10676 | -0.000 | -0.1044 | No | ||
| 113 | NFKBIZ | NFKBIZ | 10728 | -0.000 | -0.1067 | No | ||
| 114 | PRNP | PRNP | 10742 | -0.000 | -0.1073 | No | ||
| 115 | TNFRSF10B | TNFRSF10B | 10856 | -0.001 | -0.1126 | No | ||
| 116 | G0S2 | G0S2 | 11140 | -0.002 | -0.1257 | No | ||
| 117 | ZNF862 | ZNF862 | 11180 | -0.002 | -0.1273 | No | ||
| 118 | C7orf49 | C7orf49 | 11287 | -0.002 | -0.1321 | No | ||
| 119 | SLC6A15 | SLC6A15 | 11397 | -0.003 | -0.1370 | No | ||
| 120 | RUNX1 | RUNX1 | 11533 | -0.003 | -0.1430 | No | ||
| 121 | MBNL2 | MBNL2 | 11583 | -0.003 | -0.1450 | No | ||
| 122 | MDH1 | MDH1 | 11609 | -0.003 | -0.1459 | No | ||
| 123 | LAMA3 | LAMA3 | 11880 | -0.004 | -0.1581 | No | ||
| 124 | HPSE | HPSE | 11998 | -0.005 | -0.1632 | No | ||
| 125 | PIK3CD | PIK3CD | 12129 | -0.005 | -0.1688 | No | ||
| 126 | DLST | DLST | 12217 | -0.005 | -0.1724 | No | ||
| 127 | LRRFIP1 | LRRFIP1 | 12234 | -0.005 | -0.1726 | No | ||
| 128 | CXCL3 | CXCL3 | 12327 | -0.006 | -0.1764 | No | ||
| 129 | ADAM8 | ADAM8 | 12370 | -0.006 | -0.1778 | No | ||
| 130 | KANK4 | KANK4 | 12488 | -0.006 | -0.1827 | No | ||
| 131 | MRGPRX3 | MRGPRX3 | 12492 | -0.006 | -0.1822 | No | ||
| 132 | ULBP2 | ULBP2 | 12567 | -0.007 | -0.1851 | No | ||
| 133 | CCL20 | CCL20 | 12611 | -0.007 | -0.1864 | No | ||
| 134 | LRRC8C | LRRC8C | 12614 | -0.007 | -0.1859 | No | ||
| 135 | PHACTR4 | PHACTR4 | 12711 | -0.007 | -0.1897 | No | ||
| 136 | DLL1 | DLL1 | 12760 | -0.007 | -0.1913 | No | ||
| 137 | FGFR2 | FGFR2 | 12843 | -0.008 | -0.1944 | No | ||
| 138 | LATS1 | LATS1 | 12911 | -0.008 | -0.1968 | No | ||
| 139 | CXCL5 | CXCL5 | 12926 | -0.008 | -0.1968 | No | ||
| 140 | PTGS2 | PTGS2 | 13295 | -0.009 | -0.2131 | No | ||
| 141 | PPBP | PPBP | 13429 | -0.010 | -0.2185 | No | ||
| 142 | ZNF273 | ZNF273 | 13509 | -0.010 | -0.2212 | No | ||
| 143 | FERMT1 | FERMT1 | 13596 | -0.010 | -0.2243 | No | ||
| 144 | GRHL2 | GRHL2 | 13723 | -0.011 | -0.2292 | No | ||
| 145 | ARG2 | ARG2 | 13796 | -0.011 | -0.2315 | No | ||
| 146 | PI3 | PI3 | 13838 | -0.011 | -0.2324 | No | ||
| 147 | TNRC6B | TNRC6B | 13850 | -0.011 | -0.2318 | No | ||
| 148 | DUSP6 | DUSP6 | 13939 | -0.012 | -0.2348 | No | ||
| 149 | TMEM45B | TMEM45B | 13977 | -0.012 | -0.2355 | No | ||
| 150 | EGR1 | EGR1 | 13994 | -0.012 | -0.2351 | No | ||
| 151 | MFAP2 | MFAP2 | 14033 | -0.012 | -0.2358 | No | ||
| 152 | MTUS1 | MTUS1 | 14054 | -0.012 | -0.2356 | No | ||
| 153 | GJB5 | GJB5 | 14317 | -0.013 | -0.2466 | No | ||
| 154 | DENND2C | DENND2C | 14350 | -0.013 | -0.2469 | No | ||
| 155 | SERPINB5 | SERPINB5 | 14570 | -0.014 | -0.2558 | No | ||
| 156 | RAPH1 | RAPH1 | 14606 | -0.014 | -0.2561 | No | ||
| 157 | FAM46B | FAM46B | 14624 | -0.014 | -0.2556 | No | ||
| 158 | CD274 | CD274 | 14837 | -0.015 | -0.2641 | No | ||
| 159 | CD55 | CD55 | 15072 | -0.016 | -0.2735 | No | ||
| 160 | ARHGAP25 | ARHGAP25 | 15330 | -0.017 | -0.2839 | No | ||
| 161 | MXD1 | MXD1 | 15619 | -0.019 | -0.2957 | No | ||
| 162 | FAM110C | FAM110C | 15623 | -0.019 | -0.2941 | No | ||
| 163 | FGFBP1 | FGFBP1 | 15926 | -0.020 | -0.3064 | No | ||
| 164 | FAM83A | FAM83A | 16193 | -0.021 | -0.3169 | No | ||
| 165 | HMGN3 | HMGN3 | 16251 | -0.022 | -0.3175 | No | ||
| 166 | COL12A1 | COL12A1 | 16288 | -0.022 | -0.3172 | No | ||
| 167 | B3GNT5 | B3GNT5 | 16407 | -0.022 | -0.3206 | No | ||
| 168 | INPP1 | INPP1 | 16488 | -0.023 | -0.3223 | No | ||
| 169 | MAP1LC3B | MAP1LC3B | 16801 | -0.024 | -0.3346 | No | ||
| 170 | TNS4 | TNS4 | 16815 | -0.024 | -0.3330 | No | ||
| 171 | FLRT3 | FLRT3 | 16911 | -0.025 | -0.3351 | No | ||
| 172 | TCF7L2 | TCF7L2 | 16950 | -0.025 | -0.3346 | No | ||
| 173 | ODC1 | ODC1 | 17173 | -0.026 | -0.3426 | No | ||
| 174 | CHST11 | CHST11 | 17182 | -0.026 | -0.3405 | No | ||
| 175 | EREG | EREG | 17229 | -0.027 | -0.3402 | No | ||
| 176 | LPAR2 | LPAR2 | 17240 | -0.027 | -0.3382 | No | ||
| 177 | ARHGAP27 | ARHGAP27 | 17251 | -0.027 | -0.3362 | No | ||
| 178 | PNPLA8 | PNPLA8 | 17433 | -0.028 | -0.3421 | No | ||
| 179 | MALL | MALL | 17453 | -0.028 | -0.3404 | No | ||
| 180 | JHDM1D | JHDM1D | 17469 | -0.028 | -0.3385 | No | ||
| 181 | FBXO9 | FBXO9 | 17493 | -0.028 | -0.3370 | No | ||
| 182 | MCL1 | MCL1 | 17541 | -0.028 | -0.3366 | No | ||
| 183 | ADAMTS15 | ADAMTS15 | 17590 | -0.029 | -0.3362 | No | ||
| 184 | CASP1 | CASP1 | 17665 | -0.029 | -0.3370 | No | ||
| 185 | SEMA4C | SEMA4C | 17714 | -0.029 | -0.3365 | No | ||
| 186 | SH2D5 | SH2D5 | 17771 | -0.029 | -0.3364 | No | ||
| 187 | SMTN | SMTN | 17798 | -0.030 | -0.3348 | No | ||
| 188 | CXCL2 | CXCL2 | 17851 | -0.030 | -0.3345 | No | ||
| 189 | SRRM2 | SRRM2 | 17926 | -0.030 | -0.3351 | No | ||
| 190 | C19orf10 | C19orf10 | 17951 | -0.031 | -0.3334 | No | ||
| 191 | EHD1 | EHD1 | 18030 | -0.031 | -0.3342 | No | ||
| 192 | CYP27B1 | CYP27B1 | 18214 | -0.032 | -0.3398 | No | ||
| 193 | TGFB2 | TGFB2 | 18238 | -0.033 | -0.3378 | No | ||
| 194 | PHLDA1 | PHLDA1 | 18279 | -0.033 | -0.3366 | No | ||
| 195 | CYP2R1 | CYP2R1 | 18322 | -0.033 | -0.3355 | No | ||
| 196 | PTPRE | PTPRE | 18369 | -0.033 | -0.3346 | No | ||
| 197 | PLXNA2 | PLXNA2 | 18553 | -0.035 | -0.3399 | No | ||
| 198 | CALU | CALU | 18621 | -0.035 | -0.3398 | No | ||
| 199 | IL13RA2 | IL13RA2 | 18711 | -0.036 | -0.3406 | No | ||
| 200 | SDC1 | SDC1 | 18831 | -0.037 | -0.3427 | No | ||
| 201 | BMP2 | BMP2 | 18843 | -0.037 | -0.3398 | No | ||
| 202 | UAP1 | UAP1 | 18886 | -0.038 | -0.3382 | No | ||
| 203 | STX1A | STX1A | 19143 | -0.040 | -0.3465 | No | ||
| 204 | MAP7D1 | MAP7D1 | 19340 | -0.042 | -0.3518 | No | ||
| 205 | PLAUR | PLAUR | 19606 | -0.045 | -0.3601 | No | ||
| 206 | LHFPL2 | LHFPL2 | 19740 | -0.046 | -0.3620 | No | ||
| 207 | GK | GK | 20062 | -0.051 | -0.3724 | No | ||
| 208 | CXCL1 | CXCL1 | 20079 | -0.051 | -0.3684 | No | ||
| 209 | TRIM22 | TRIM22 | 20318 | -0.055 | -0.3744 | Yes | ||
| 210 | TSC22D1 | TSC22D1 | 20383 | -0.056 | -0.3722 | Yes | ||
| 211 | GAS1 | GAS1 | 20486 | -0.058 | -0.3716 | Yes | ||
| 212 | CYP1B1 | CYP1B1 | 20518 | -0.059 | -0.3677 | Yes | ||
| 213 | SKP2 | SKP2 | 20596 | -0.060 | -0.3657 | Yes | ||
| 214 | TFPI2 | TFPI2 | 20632 | -0.062 | -0.3616 | Yes | ||
| 215 | B4GALT1 | B4GALT1 | 20633 | -0.062 | -0.3559 | Yes | ||
| 216 | PPIF | PPIF | 20717 | -0.064 | -0.3538 | Yes | ||
| 217 | SLC25A37 | SLC25A37 | 20764 | -0.065 | -0.3499 | Yes | ||
| 218 | TUBA4A | TUBA4A | 20809 | -0.067 | -0.3458 | Yes | ||
| 219 | S100A6 | S100A6 | 20850 | -0.068 | -0.3414 | Yes | ||
| 220 | ARNTL2 | ARNTL2 | 20855 | -0.068 | -0.3353 | Yes | ||
| 221 | ATP2B1 | ATP2B1 | 20901 | -0.069 | -0.3310 | Yes | ||
| 222 | MMP14 | MMP14 | 20926 | -0.070 | -0.3256 | Yes | ||
| 223 | TIMP1 | TIMP1 | 21029 | -0.074 | -0.3235 | Yes | ||
| 224 | LRP8 | LRP8 | 21057 | -0.075 | -0.3178 | Yes | ||
| 225 | GTPBP2 | GTPBP2 | 21111 | -0.078 | -0.3131 | Yes | ||
| 226 | PHLDA2 | PHLDA2 | 21151 | -0.080 | -0.3075 | Yes | ||
| 227 | SLC20A1 | SLC20A1 | 21191 | -0.083 | -0.3017 | Yes | ||
| 228 | SLC9A1 | SLC9A1 | 21248 | -0.087 | -0.2963 | Yes | ||
| 229 | ITPRIP | ITPRIP | 21293 | -0.090 | -0.2900 | Yes | ||
| 230 | EPHA2 | EPHA2 | 21313 | -0.092 | -0.2823 | Yes | ||
| 231 | HBEGF | HBEGF | 21345 | -0.097 | -0.2748 | Yes | ||
| 232 | LIF | LIF | 21404 | -0.105 | -0.2677 | Yes | ||
| 233 | LDLR | LDLR | 21410 | -0.105 | -0.2582 | Yes | ||
| 234 | SESN2 | SESN2 | 21424 | -0.107 | -0.2489 | Yes | ||
| 235 | NT5E | NT5E | 21448 | -0.112 | -0.2395 | Yes | ||
| 236 | IL1A | IL1A | 21453 | -0.113 | -0.2292 | Yes | ||
| 237 | IER3 | IER3 | 21480 | -0.122 | -0.2191 | Yes | ||
| 238 | SDC4 | SDC4 | 21489 | -0.125 | -0.2079 | Yes | ||
| 239 | IL1B | IL1B | 21493 | -0.129 | -0.1961 | Yes | ||
| 240 | CLCF1 | CLCF1 | 21517 | -0.146 | -0.1837 | Yes | ||
| 241 | AHNAK2 | AHNAK2 | 21523 | -0.148 | -0.1702 | Yes | ||
| 242 | NDRG1 | NDRG1 | 21524 | -0.149 | -0.1563 | Yes | ||
| 243 | TNFRSF12A | TNFRSF12A | 21534 | -0.154 | -0.1425 | Yes | ||
| 244 | FOSL1 | FOSL1 | 21546 | -0.167 | -0.1275 | Yes | ||
| 245 | TRIB1 | TRIB1 | 21555 | -0.173 | -0.1118 | Yes | ||
| 246 | PTHLH | PTHLH | 21558 | -0.176 | -0.0955 | Yes | ||
| 247 | IL11 | IL11 | 21567 | -0.185 | -0.0787 | Yes | ||
| 248 | AGPAT9 | AGPAT9 | 21568 | -0.188 | -0.0613 | Yes | ||
| 249 | TMEM154 | TMEM154 | 21569 | -0.190 | -0.0437 | Yes | ||
| 250 | DUSP5 | DUSP5 | 21581 | -0.219 | -0.0238 | Yes | ||
| 251 | GDF15 | GDF15 | 21592 | -0.263 | 0.0001 | Yes |

